Abstract
Lactic acid bacteria (LAB) confer health benefits to human when administered orally. We have recently isolated several species of LAB strains from plant sources, such as fruits, vegetables, flowers, and medicinal plants. Since antibiotics used to treat bacterial infection diseases induce the emergence of drug-resistant bacteria in intestinal microflora, it is important to evaluate the susceptibility of LAB strains to antibiotics to ensure the safety and security of processed foods. The aim of the present study is to determine the minimum inhibitory concentration (MIC) of antibiotics against several plant-derived LAB strains. When aminoglycoside antibiotics, such as streptomycin (SM), kanamycin (KM), and gentamicin (GM), were evaluated using LAB susceptibility test medium (LSM), the MIC was higher than when using Mueller–Hinton (MH) medium. Etest, which is an antibiotic susceptibility assay method consisting of a predefined gradient of antibiotic concentrations on a plastic strip, is used to determine the MIC of antibiotics world-wide. In the present study, we demonstrated that Etest was particularly valuable while testing LAB strains. We also show that the low susceptibility of the plant-derived LAB strains against each antibiotic tested is due to intrinsic resistance and not acquired resistance. This finding is based on the whole-genome sequence information reflecting the horizontal spread of the drug-resistance genes in the LAB strains.
Similar content being viewed by others
Introduction
Lactic acid bacteria (LAB), which consist of over 10 genera including Enterococcus, Lactobacillus, Lactococcus, Streptococcus, Pediococcus, and Weissella [1,2,3], contribute to promote human health [4, 5]. “Probiotics” are the living microorganisms that contribute health benefits to the host when administered orally in appropriate amounts [6]. LAB, which are typical probiotics, have been used to produce fermented foods, beverages, and supplements that claim some health benefits [7,8,9,10].
The LAB strains are roughly classified into two groups by the isolated environment: one is isolated from animal sources (designated as animal-derived LAB), such as skin and intestine. Another is isolated from plant sources (designated as plant-derived LAB), such as fruits, vegetables, and medicinal plants. The animal-derived LAB strains have been widely used to manufacture yogurt and cheese preparation, whereas the plant-derived ones have been useful to produce fermented dishes, mainly in East- and Southeast-Asians. We have isolated many kinds of plant-derived LAB strains from vegetables, flowers, fruits, and medicinal herbs. In a plant-derived LAB library composed of over 700 strains, we have found that several LAB strains are very useful for immune modulation, the improvement of liver function, and the reduction of obesity [11,12,13,14]. The LAB strains we characterize in our academic laboratories could be the source of health promoting organisms for the health food industry.
Since LAB strains are often administrated orally as living organisms contained in fermented foods, and considering that the emergence of antibiotic-resistant bacteria is caused by the clinical use of antibiotics (antibacterial agents), evaluation of the susceptibility of LAB strains to antibiotics is very important for food safety and security. In other words, if certain LAB strains that live in the intestinal tract are exposed to antibiotics, they may acquire resistance to antibiotics by the horizontal transfer of the drug-resistance gene. The risk of horizontal gene transfer is higher than when the intrinsic resistance gene is encoded on the chromosome if the resistance gene is mediated by a plasmid or transposon [15, 16].
In this study, we evaluated the antibiotic susceptibilities of plant-derived LAB isolates that have been previously confirmed to exhibit the health-promoting effects. Further, the risks of horizontal spread were evaluated through whole-genome sequence analysis of the strains.
Materials and methods
Bacterial strains, growth media, and culture conditions
The antibiotic susceptibility of six strains of the plant-derived LAB was investigated. Table 1 shows the LAB strains isolated previously by our research group [11,12,13,14, 17,18,19,20,21]. To culture LAB strains, MRS medium (Becton, Dickinson, and Company, Franklin Lakes, NJ, USA) was used. To evaluate the MIC, the LAB strains were cultured in MH medium (Becton, Dickinson, and Company) or LSM medium [22] composed of 90% Iso-Sensitest broth [23] (Oxoid, Cheshire, UK) and 10% (w/v) MRS medium. If necessary, 1.5 (w/v) % agar was added to the medium.
Evaluation of susceptibility to antibiotics using the broth microdilution method
Broth microdilution test method was performed using a U-bottom 96-well microtiter plate. Antibiotics evaluated in the study were tetracycline (TC), erythromycin (EM), chloramphenicol (CL), ampicillin (AM), vancomycin (VA), gentamicin (GM), clindamycin (CM), kanamycin (KM), streptomycin (SM), and tylosin (TS), which were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). Each antibiotic was diluted into LSM or MH medium under the appropriate concentration. A 200 μl portion of the LSM or MH medium was transferred into each well to contain given concentrations (serially diluted) of antibiotics. After inoculation of LAB cells at 5 × 105 colony-forming unit (CFU) ml−1, the plate was incubated at 28 or 37 °C until cell growth was observed in the control well containing medium without antibiotics. After cultivation, the bacterial growth measured as turbidity or cell density in the well bottom. The MIC is defined as the lowest concentration of the added antibiotic at which growth of the plant-derived LAB strain is completely inhibited or at which small button of bacteria (no more than 2 mm in diameter) was only observed [24, 25]. This examination was repeated at least three times, and the average value was recorded.
Evaluation of MIC using Etest strips
MIC measurement using the Etest strip method (bioMérieux, Lyon, France) was performed according to the manufacturer’s instructions: LAB cells grown until the stationary phase were suspended in a brain heart infusion (BHI) medium. The cell suspension (5 × 108 CFU ml−1) soaked in a sterile cotton swab was streaked with a plate rotation 60 degrees three times on the LSM agar medium. After drying the surface of plate, Etest strips were applied to the agar medium, followed by incubation at 28, 37, or 45 °C until cell growth was observed on the control agar plate. The experiment was repeated at least three times, and the average value was recorded.
Whole-genome sequencing and annotation
The chromosomal DNA from each LAB strain was isolated, as described previously [20]. The whole-genome sequencing of G-15, 15-1A, and SN13T strains was done on a next-generation sequencing platform, PacBio RS II (Pacific Biosciences, Menlo Park, CA, U.S.A.), as performed in previous studies [19, 20]. After de novo assembling, the obtained genomic contig sequence data was annotated using the Microbial Genome Annotation Pipeline (MiGAP) [26]. The genome data were analyzed using the In Silico Molecular Cloning Genomic Edition (In Silico Biology, Inc., Yokohama, Japan).
Phylogenetic analysis
The predicted proteins that have the motifs of serine hydrolase and metal-dependent hydrolase coded on G-15 chromosomal DNA (G15_0480, G15_0661, G15_0663, G15_0941, G15_0720, G15_0818, G15_0918, G15_1504, G15_1771, and G15_2962) were phylogenetically analyzed with typical class A–D β-lactamases as follows: class A enzymes from Escherichia (E.) coli (CU928163 and NC_019081), Klebsiella (K.) pneumoniae (AY850171), and Proteus vulgaris (D29982); Class B ones from K. pneumoniae (NC_014312) and Pseudomonas (P.) aeruginosa (NC_022345); Class C ones from E. coli (HQ185697), K. pneumoniae (AF259520), and P. aeruginosa (AF490770); and Class D ones from Acinetobacter baumannii (NC_025109), E. coli (NC_022374), and K. pneumoniae (NC_019160).
The eighteen predicted proteins of the 15-1A strain annotated to efflux pump (EM151A_0036, EM151A_0081, EM151A_0695, EM151A_1022, EM151A_1101, EM151A_1104, EM151A_1120, EM151A_1681, EM151A_1744, EM151A_1998, EM151A_2046, EM151A_2214, EM151A_2250, EM151A_2303, EM151A_2455, EM151A_2653, EM151A_2698, and EM151A_3018) were also analyzed with MsrA proteins from S. dysgalactiae subsp. equisimilis (AP011114) and Streptococcus sp. (DQ131177) and MefA proteins from S. pyogenes (KJ809088) and S. viridans (EF042094).
The phylogenetic trees were drawn with the ClustalW program in Molecular Evolutionary Genetics Analysis (MEGA) software ver. 6.0 [27] using the unweighted pair group method with arithmetic (UPGMA) [28]. The bootstrap values [29] were determined from 5 000 replications.
Acute oral toxicity and mutagenicity tests
The acute oral toxicity using rats of each LAB strain, 15-1A, SN13T and LP28, was evaluated by the New Drug Development Research Center, Inc., and the Japan Food Research Laboratories (for the IJH-SONE68 strain), as described in our previous studies [17, 19]. The test was performed according to the OECD Guidelines for the Testing of Chemicals, Guideline 420 (2001).
The mutagenicity test (umu test) of the culture supernatant of the LAB strain was performed using a Umulac AT test kit (Protein Purify Ltd., Maebashi, Japan) according to the manufacturer’s protocol. The induction of the umu gene is responsible for DNA damage that was calculated by the umuC-lacZ fusion gene expression in Salmonella enterica serovar Typhimurium NM2009. If the sample added into the reaction mixture enhances the activity of β-galactosidase to more than twice that of background reaction, it is speculated that the sample exhibits the mutagenic property at the given concentration.
Results
Susceptibility to antibiotics as measured by the broth microdilution method
The MICs of antibiotics measured by the broth microdilution method with the LSM are shown in Table 2. The MIC (16 ≤ μg ml−1) of clindamycin (CM) on Enterococcus (E.) mundtii 15-1A isolated from Brassica rapa L. subsp. nipposinica (L.H. Bailey) Hanelt var. linearifolia [18] was higher than the cutoff value (4-μg ml−1). However, the MIC of CM on other LAB isolates was lower than the cutoff value. In addition, among the MICs of ampicillin (AM), kanamycin (KM), streptomycin (SM), and gentamicin (GM) on the LAB strains, some strains had higher MICs than the cutoff values defined by the European Food Safety Authority (EFSA). The EFSA provides independent scientific advice on risks in the food chain. The MICs of AM, KM, SM, and GM were also evaluated using MH medium, designed to test the sensitivity of pathogens to antibiotics [30], and not for LSM. As shown in Table 3, when using MH medium, the MIC of each antibiotic was eight times lower than with the LSM. The MICs of other antibiotics except AM against Enterococcus (E.) avium G-15 were less than or equal to the cutoff values.
Antibiotic MIC determination using Etest strips
The MICs of tested antibiotics were also measured using Etest strips located on the LSM agar. As shown in Table 4, almost all MICs were lower or higher than those determined by the broth microdilution method using LSM or MH medium, respectively. With the Etest strip, the MIC of AM against E. avium G-15 and Lactobacillus (Lb.) plantarum SN13T were higher than the cutoff values. Further, the MIC of SM on Pediococcus (P.) pentosaceus LP28 was also higher than the cutoff value.
Whole-genome sequence analyses
Each whole-genome sequence of E. avium G-15, E. mundtii 15-1A, and Lb. plantarum SN13T was determined. These strains have been isolated from a carrot, Citras Iyo, and a pork sausage fermented by wrapping it with a banana leaf, called “nem chua,” respectively. The whole-DNA sequences are summarized in Table 1, together with those of the fig leaf–derived Lb. paracasei IJH-SONE68, Lb. plantarum SN35N, and P. pentosaceus LP28 determined previously [19,20,21]. The whole-genome sequences of the G-15, 15-1A, and SN13T strains reveal that the three whole genomes are circular DNA, and the sizes are 3,623,727, 3,112,343, and 3,612,790 bp, with GC contents of 39.7%, 38.5%, and 46.4%, respectively. To obtain the GenBank/EMBL/DDBJ accession numbers, the nucleotide sequence data of the G-15, 15-1A, and SN13T strains have been deposited in the International Nucleotide Sequence Database (AP019814, AP019810, and AP019815, respectively). The average genome sizes of other Lb. plantarum and E. mundtii strains, which were calculated from the completely sequenced genomic DNA data registered in the NCBI genome database, were 3318 (from 2952 to 3697; n = 90) and 3 174 (from 2827 to 3505; n = 4) kb, respectively. However, at this time, the complete genomic sequence of E. avium has not been previously registered in the NCBI database.
From the whole-genome sequence data, it was found that E. mundtii 15-1A harbors three kinds of plasmids, which were designated pEM15-1A-1, 2, and 3 (AP019811, AP019812, and AP019813, respectively). The sizes of pEM15-1A-1, pEM15-1A-2, and pEM15-1A-3 are 67,440, 59,372, and 56,627 bp, respectively. The GC content of each plasmid is 36.1%, 33.9%, and 35.7%, respectively. Lb. plantarum SN13T harbors a plasmid designated pSN13T-1 (AP019816), with a size of 72,292 bp and a GC content of 40.4%. Significantly, E. avium G-15 did not have any plasmids.
Safety evaluations of the plant-derived LAB strains
We evaluated the acute toxicity of the E. mundtii 15-1A, Lb. paracasei IJH-SONE68, Lb. plantarum SN13T, and P. pentosaceus LP28 cells using rats, confirming that the ingestion of each strain did not induce illness or death. Additionally, no differences were noted in the health of the animals and no signs of inflammation in the organs were observed by histology. Furthermore, the umu test demonstrates that no culture supernatant of any strain induces mutagenesis. We have previously determined that the G-15 and SN35N cells and the culture supernatant have no acute toxicity or mutagenic activity, respectively [17, 20].
Discussion
In general, LAB require a nutrient-rich medium containing carbohydrates, vitamins, amino acids, and minerals for growth. In order to satisfy the nutrition requirements, de Man, Rogosa, and Sharpe (MRS) medium has been developed [31]. Although some nutrients contained in the MRS medium could be antagonistic to antibiotic, but the mechanism is not fully understood [22, 32, 33]. To avoid antibiotic-inhibitory activity by different medium constituents, MH medium was used to determine the MIC values. However, the MH medium appears to be inadequate for cultivating LAB. In fact, our isolates of Lb. reuteri, Lb. sakei, and Lb. amylovorous grew very poorly in MH medium (data not shown). In the present study, we evaluated the susceptibility of health-promoting LAB strains against several antibiotics using LSM, which was designed as an alternative to the MH medium for LAB [22].
The MICs of AM and aminoglycosides (SM, KM, GM), which were determined using the broth microdilution method with LSM, were higher than those with MH medium, as shown in Tables 2 and 3, respectively. The MICs of GM on Staphylococcus aureus, E. faecalis, and E. coli, which have been determined using the broth microdilution method with the LSM, were also higher than those with the MH one [22]. Judging from the report by another research group, some compounds in LSM are likely to interfere with the antimicrobial activity of some antibiotics.
Interestingly, when Etest strip was employed, almost all of the MICs were lower than those determined using the broth microdilution method, especially for aminoglycosides (Table 4). Furthermore, the low sensitivity of E. avium G-15 to AM observed in the assay using the MH medium was also observed in the Etest assay. The Etest strip method has been reported to be an applicable technique for evaluating the MICs of Gram-positive non-spore-forming anaerobes, including bifidobacteria and some LAB strains [34,35,36]. However, when the MH agar medium was used instead of the LSM agar one for the Etest assay, the inhibitory zone was very unclear, suggesting that the MH medium is not suitable (data not shown). Therefore, the Etest assay with the LSM may be useful to evaluate the MICs on LAB.
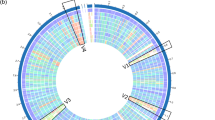
β-Lactamases (EC3.5.2.6), which are produced by bacteria and confer resistance to β-lactam antibiotics, are classified into four groups (Classes A, B, C, and D): penicillinase and cephalosporinase belong to Class A and C, respectively. Oxacillinase (Class D) belongs to the serine hydrolase family enzyme. The Class B enzyme carbapenemase is a member of metal-dependent hydrolase [37]. The whole-genome sequence of E. avium G-15 suggests that there are six (G15_0480, G15_0661, G15_0663, G15_0941, G15_1504, and G15_2962) and four (G15_0720, G15_0818, G15_0918, and G15_1771) genes encoding proteins that have the motifs of serine hydrolase and metal-dependent hydrolase on the chromosomal DNA, respectively. The 10 predicted proteins were compared with 12 typical enzymes of Class A–D β-lactamases by phylogenic tree analysis, showing that not all of the predicted hydrolases are classified as β-lactamases (Fig. 1a). It has been reported that the low sensitivity or resistance of Enterococcus species to β-lactams is a genus-specific feature caused by low affinity of their penicillin-binding proteins [38, 39]. In addition, Murray reported in her review that the MICs of AM and penicillin for E. faecalis strains are 1–4 and 2–8 μg ml−1, respectively, and those values are 10–100 times higher than those of most streptococci [38]. Therefore, the MIC of AM on E. avium G-15, which has been shown to be two times higher than the cutoff value in the MH medium, is thought to be due to intrinsic resistance rather than acquired resistance.
Phylogenetic trees on the β-lactamases a and multi-drug efflux pumps b. The phylogenetic trees were drawn with the ClustalW program in Molecular Evolutionary Genetics Analysis (MEGA) software ver. 6.0 [27] using the unweighted pair group method with arithmetic (UPGMA) [28]. The bootstrap values [29] were determined from 5 000 replications. The bottom horizontal bars show a distance of 0.1 substitutions per site
The MIC of CM on E. mundtii 15-1A was 16 times higher than the cutoff value when using broth microdilution method, although the resistance was not observed when using the Etest method. Mutations in ribosomal proteins L4 and L22 (K63E and Δ82–84, respectively, E. coli numbering) [40, 41] and 23S rRNA (A2058 and A2059, E. coli numbering) [42] are known factors that decrease the sensitivities of bacteria to the macrolide-lincosamide-streptogramin (MLS) group of antibiotics, including EM and CM. Based on the whole-genome sequence, E. mundtii 15-1A has no mutations on corresponding residues or nucleotides in deduced ribosomal proteins (encoded by EM151A_1637 and EM151A_1641) or 23S rRNA sequences, respectively.
Furthermore, multi-drug efflux pumps have been reported to contribute to antibiotic resistance in bacteria. Some pathogenic Streptococcus spp. have efflux pumps named MsrA and MefA that give the bacteria an acquired resistance against MLS antibiotics [43]. Among the predicted CDSs (coding sequences) in E. mundtii 15-1A, there are 18 gene products annotated to efflux pump. However, all of the predicted pumps were found to be clearly different from MsrA and MefA via phylogenic tree analysis of those proteins (Fig. 1b). Therefore, those predicted genes do not seem to play a role in acquired MLS antibiotic resistance. It has also been reported that some E. mundtii isolates have low sensitivities against lincosamides without known resistance genes, such as ermA, ermB, ermC, mefA, and msrA [44]. In fact, the PCR analyses performed on E. mundtii 15-1A using primer sets designed for amplifying those five genes [45,46,47] indicate that the 15-1A strain does not have those resistance genes (data not shown). Thus, the observed higher MICs of MLSs on E. mundtii 15-1A are thought to be not acquired but intrinsic characteristics. The accumulation of genomic and strain characteristics of minor enterococci, including E. avium and E. mundtii, will provide detailed information on the differences in susceptibility against antibiotics within the genus.
Although Streptococcus (S.) thermophilus has been widely and traditionally used to manufacture yogurt [48], the genus Streptococcus includes some harmful species, such as S. agalactiae, S. pneumoniae, and S. pyogenes. Bolotin et al. reported that all streptococci lack the gene encoding RecQ helicase [49], which is present in bacteria from mammals and increases genome stability [47]. Furthermore, the pathogenic streptococci, but not S. thermophilus, lack the sbcC and sbcD genes that contribute to genome stabilization by repairing the recombinogenic double-strand DNA brakes, suggesting that genomic instability might allow pathogenic streptococci to acquire virulence-related genes [49, 50]. Whole-genome sequencing revealed that the six strains analyzed in the present study have all three of those genes (recQ, sbcC, and sbcD). The result indicates that our isolates are genomically stable and resistant to undesirable genes that bring antibiotic resistance and pathogenicity via horizontal gene transfer. In addition, it has been shown that the isolates do not cause disease or mutagenesis after administration, suggesting that those isolates are useful for the production of fermented foods and healthcare materials.
References
Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–96.
Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71.
Fusco V, Quero GM, Cho GS, Kabisch J, Meske D, Neve H, et al. The genus Weissella: taxonomy, ecology and biotechnological potential. Front Microbiol. 2015;6:155.
Meydani SN, Ha WK. Immunologic effects of yogurt. Am J Clin Nutr. 2000;71:861–72.
Adolfsson O, Meydani SN, Russell RM. Yogurt and gut function. Am J Clin Nutr. 2004;80:245–56.
Sanders ME. Probiotics: definition, source, selection, and uses. Clin Infect Dis. 2008;46(Suppl. 2):S58–61. discussion, S144–51.
Kleerebezem M, de Vos WM. Lactic acid bacteria: life after genomics. Micro Biotechnol. 2011;4:318–22.
Evivie SE, Huo GC, Igene JO, Bian X. Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr Res. 2017;61:1318034.
West CE, Dzidic M, Prescott SL, Jenmalm MC. Bugging allergy; role of pre-, pro- and synbiotics in allergy prevention. Allergol Int. 2017;66:529–38.
Mahasneh SA, Mahasneh AM. Probiotics: a promising role in dental health. Dent J (Basel). 2017;5:26.
Jin H, et al. Establishment of an in vitro Peyer’s patch cell culture system correlative to in vivo study using intestine and screening of lactic acid bacteria enhancing intestinal immunity. Biol Pharm Bull. 2010;33:289–93.
Higashikawa F, et al. Improvement of constipation and liver function by plant-derived lactic acid bacteria: a double-blind, randomized trial. Nutrition. 2010;26:367–74.
Zhao X, et al. The obesity and fatty liver are reduced by plant-derived Pediococcus pentosaceus LP28 in high fat diet-induced obese mice. PLoS ONE. 2012;7:e30696.
Higashikawa F, et al. Antiobesity effect of Pediococcus pentosaceus LP28 on overweight subjects: a randomized, double-blind, placebo-controlled clinical trial. Eur J Clin Nutr. 2016;70:582–7.
Devirgiliis C, Barile S, Perozzi G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011;6:275–84.
van Reenen CA, Dicks LM. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: what are the possibilities? A review. Arch Microbiol. 2011;193:157–68.
Tamura T, et al. Establishment of an efficient fermentation system of gamma-aminobutyric acid by a lactic acid bacterium, Enterococcus avium G-15, isolated from carrot leaves. Biol Pharm Bull. 2010;33:1673–9.
Jeon HJ, Noda M, Matoba Y, Kumagai T, Sugiyama M. Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im. Biochem Biophys Res Commun. 2009;378:574–8.
Noda M, et al. A novel structure of exopolysaccharide produced by a plant-derived lactic acid bacterium Lactobacillus paracasei IJH-SONE68. J Biochem. 2018;164:87–92.
Noda M, Shiraga M, Kumagai T, Danshiitsoodol N, Sugiyama M. Characterization of the SN35N strain-specific exopolysaccharide encoded in the whole circular genome of a plant-derived Lactobacillus plantarum. Biol Pharm Bull. 2018;41:536–45.
Yasutake T, et al. Characterization of the LP28 strain-specific exopolysaccharide biosynthetic gene cluster found in the whole circular genome of Pediococcus pentosaceus. Biochem Biophys Rep. 2016;5:266–71.
Klare I, et al. Evaluation of new broth media for microdilution antibiotic susceptibility testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl Environ Microbiol. 2005;71:8982–6.
Turnidge JD, Bell JM. Antimicrobial Susceptibility on Solid Media. In: Lorian V, editor. Antibiotics in Laboratory Medicine. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2005.
Clinical and Laboratory Standards Institute, Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard – Ninth edition, CLSI DocumentM07-A9. 2012.
Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. CLSI Document M100-S22. 2012.
Sugawara H, Ohyama A, Mori H, Kurokawa K Microbial Genome Annotation Pipeline (MiGAP) for diverse users. S001-1-2. The 20th International Conference on Genome Informatics (GIW2009) Poster and Software Demonstrations, Yokohama, Japan. 2009.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;38:1409–38.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Mueller HJ, Hinton J. A protein-free medium for primary isolation of the Gonococcus and Meningococcus. Proc Soc Expt Biol Med. 1941;48:330–3.
de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;23:130–5.
Danielsen M, Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int J Food Microbiol. 2003;82:1–11.
Huys G, D’Haene K, Swings J. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett Appl Microbiol. 2002;34:402–6.
Blandino G, Milazzo I, Fazio D. Antibiotic susceptibility of bacterial isolates from probiotic products available in Italy. Micro Ecol Health Dis. 2008;20:199–203.
Mättö J, Suihko ML, Saarela M. Comparison of thre Etest media for antimicrobial susceptibility testing of bifidobacteria using the Etest method. Int J Antimicrob Agents. 2006;28:42–8.
Citron DM, Ostovari MI, Karlsson A, Goldstein EJ. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J Clin Microbiol. 1991;29:2197–203.
Bush K. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother. 2018;62:e01076–18.
Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65.
Williamson R, le Bouguénec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985;131:1933–40.
Chittum HS, Champney WS. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–8.
Lovmar M, et al. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. EMBO J. 2009;28:736–44.
Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12.
Singh KV, Malathum K, Murray BE. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob Agents Chemother. 2001;45:263–6.
Loch IM, Glenn K, Zadoks RN. Macrolide and lincosamide resistance genes of environmental streptococci from bovine milk. Vet Microbiol. 2005;111:133–8.
Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–6.
Clancy J, et al. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–79.
Wondrack L, Massa M, Yang BV, Sutcliffe J. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother. 1996;40:992–8.
Bourlioux P, Pochart P. Nutritional and health properties of yogurt. World Rev Nutr Diet. 1988;56:217–58.
Bolotin A, et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotech. 2004;22:1554–8.
Bidnenko V, et al. sbcB sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol Microbiol. 1999;33:846–57.
Acknowledgements
We thank the Analysis Center of Life Science, Hiroshima University, for the use of their facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Noda, M., Danshiitsoodol, N., Inoue, Y. et al. Antibiotic susceptibility of plant-derived lactic acid bacteria conferring health benefits to human. J Antibiot 72, 834–842 (2019). https://doi.org/10.1038/s41429-019-0218-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0218-4
This article is cited by
-
Phosphate solubilization and plant growth properties are promoted by a lactic acid bacterium in calcareous soil
Applied Microbiology and Biotechnology (2024)
-
Functional probiotics of lactic acid bacteria from Hu sheep milk
BMC Microbiology (2020)
-
Rapid detection of antibiotic resistance genes in lactic acid bacteria using PMMA-based microreactor arrays
Applied Microbiology and Biotechnology (2020)