Abstract
CRISPR systems have been proven as versatile tools for site-specific genome engineering in mammalian species. During the gene editing processes, these RNA-guide nucleases introduce DNA double strand breaks (DSBs), in which non-homologous DNA end joining (NHEJ) dominates the DNA repair pathway, limiting the efficiency of homology-directed repair (HDR), the alternative pathway essential for precise gene targeting. Multiple approaches have been developed to enhance HDR, including chemical compound or RNA interference-mediated inhibition of NHEJ factors, small molecule activation of HDR enzymes, or cell cycle timed delivery of CRISPR complex. However, these approaches face multiple challenges, yet have moderate or variable effects. Here we developed a new approach that programs both NHEJ and HDR pathways with CRISPR activation and interference (CRISPRa/i) to achieve significantly enhanced HDR efficiency of CRISPR-mediated gene editing. The manipulation of NHEJ and HDR pathway components, such as CtIP, CDK1, KU70, KU80, and LIG4, was mediated by catalytically dead guide RNAs (dgRNAs), thus relying on only a single catalytically active Cas9 to perform both CRISPRa/i and precise gene editing. While reprogramming of most DNA repair factors or their combinations tested enhanced HDR efficiency, simultaneously activating CDK1 and repressing KU80 has the strongest effect with increased HDR rate upto an order of magnitude. Doxycycline-induced dgRNA-based CRISPRa/i programming of DNA repair enzymes, as well as viral packaging enabled flexible and tunable HDR enhancement for broader applicability in mammalian cells. Our study provides an effective, flexible, and potentially safer strategy to enhance precise genome modifications, which might broadly impact human gene editing and therapy.
Similar content being viewed by others
Introduction
Organisms evolved multiple mechanisms to maintain genome integrity1,2. As the cellular genome is constantly exposed to environmental damage, multiple DNA damage repair pathways exist to protect the genome from harmful or potentially catastrophic alterations3. DNA double-strand break (DSB) repair pathways are highly conserved between eukaryotes including mammalian species4,5. Non-homologous DNA end joining (NHEJ) and homology-directed repair (HDR) are two major DNA repair pathways that can either act in concert or antagonistic manner6. HDR is a pathway which uses template DNA, such as an intact sister chromosomal copy or an exogenous donor to repair the DSBs7, thus can robustly generate perfect repair8. However, HDR efficiency depends on species, cell type, and the stage of the cell cycles8,9,10,11,12. In mammalian cells, NHEJ has been considered the major pathway to repair the DNA13, whereas HDR is more common in the Saccharomyces cerevisiae14. NHEJ is an imperfect process, which often lead to gain or loss of a few nucleotides at each end of the breakage site13,15. This character can lead to subsequent deleterious genetic alteration that results in cellular malfunctioning, cancer, or aging16. The DNA repair enzymes KU70, KU80, and Ligase IV (LIG4) play central roles in NHEJ-mediated DNA repair, where KU70 and KU80 proteins stabilize the DNA ends and make them physical proximity to facilitate end ligation performed by LIG47. On the other hand, proteins such as BRCA1/2, RAD50, RAD51, and various cell cycle regulators are directly involved in homology-directed repair (HDR)9,17, although the pathway has yet to be fully characterized.
The type II bacterial adaptive immune system, clustered regularly interspaced palindromic repeats (CRISPR)-associated protein 9 (Cas9) is a powerful genome editing tool18,19,20. The Cas9 - single guide RNA (sgRNA) complex induces site-specific DSBs, which can be repaired by either of the two main DNA repair pathways, NHEJ and HDR. The error-prone repairs by NHEJ often introduce unpredictable frame shift insertions and deletions (indels), leading to loss-of-function of target genes. In contrast, HDR can either generate perfect DNA repair or precise genome modification guided by donor templates. However, HDR is substantially less efficient compared to NHEJ21,22 in mammalian cells and most often restricted to S/G2 phase(s) of the cell cycle23. Owning to the importance of HDR in mediating precise genetic modification, extensive efforts have been made to change the balance of DNA repair pathways. However, due to the intricacy of the DNA repair pathways, the available tools to enhance HDR are still limited to a few choices with relatively small effect. Moreover, little success to date has been achieved to directly augment the HDR pathway itself. Thus, manipulation of both HDR and NHEJ using simple genetic tools might enable or strengthen a variety of genome editing applications. Recently, 14-nt or 15-nt guide RNAs have been shown to be catalytically inactive yet maintain the target-site binding capacity24,25. These catalytically dead guide RNAs (dgRNAs) thus can be utilized to modulate gene expression using a catalytically active Cas924,25. Therefore, an active Cas9 nuclease can be repurposed to simultaneously perform genome editing and regulate gene transcription using both types of gRNAs in the same cell. We thus hypothesize that dgRNAs together with the associated CRISPR activation (CRISPRa) and interference (CRISPRi) modules can be deployed to achieve HDR enhancement using a single active Cas9.
Results
To enhance HDR efficiency of CRISPR-mediated gene editing with clean genetic approaches that avoid the potential side effects from chemical compounds, we leverage a method that tunes the expression of DNA damage repair pathway components by dgRNA/active Cas9-mediated CRISPRa and CRISPRi (CRISPRa/i). We constructed Com binding loop into dgRNA scaffold26 for recruiting COM-KRAB (CK) fusion domain to repress the NHEJ-related genes, as well as MS2-binding loops into dgRNA scaffold26,27 for recruiting MCP - P65-HSF1 (MPH) fusion domain to activate HDR-related genes (Fig. 1a, b). With these two constructs we first tested them using an EGFP reporter system, and then two endogenous genes. The results showed robust activation and repression of both exogenous reporter genes and endogenous genes, where the EGFP’s mRNA level was significantly upregulated by dgGFP - MS2:MPH and repressed by dgGFP-Com:CK (Supplementary Fig. S1a–c), and the transcriptional level of ASCL1 and HBG1 were dramatically upregulated by dgRNA-MS2:MPH systems with gene-specific dgRNAs (Supplementary Fig. S1d, e). Based on the robust functions of dgRNA-MS2:MPH and dgRNA-Com:CK, we programmed the activation and repression of several key HDR and NHEJ genes, respectively. The results showed that CDK1, promotes efficient end resection by phosphorylating DSB resection nuclease28, and CtIP, is well-known enzyme promoting resection of DNA ends to single-stranded DNA (ssDNA) which is essential for HR29, transcript levels were upregulated by near three-fold, and LIG4, KU70, and KU80 were reduced by 40–50% (Supplementary Fig. S1f–j).
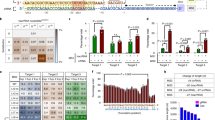
a Diagram of dgRNA-MS2:MPH expression vector for activating key genes of HDR pathway. b Diagram of dgRNA-Com:CK expression vector for repressing key genes of NHEJ pathway. c Diagram of the TLR system. Cas9/sgRNA can induce DSBs in the target site. If DSBs are repaired by NHEJ, 3n + 2 bp frame shift indels can restore mCherry expression, which accounted for approximately 1/3 mutagenic NHEJ events. Alternatively, if DSBs were repaired according an intact EGFP template, the mutations in bf-Venus will be corrected, leading to Venus (EGFP variant) expression. d Quantitative results of HDR efficiency by programming essential components of DNA repair pathways (Vector vs. dgCDK1-2, p = 0.0014; Vector vs. dgCtIP-1, p = 0.0079; Vector vs. dgLIG4-1, p = 0.0031; Vector vs. dgKU70-2, p = 0.026; Vector vs. dgKU80-1, p = 0.0338; Vector vs. dgCDK1-2 + dgCtIP-1, p = 0.0138; Vector vs. dgCDK1-2 + dgLIG4-1, p = 0.0022; Vector vs. dgCDK1-2 + dgKU70-2, p = 0.075; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.0493; Vector vs. dgCtIP-1 + dgLIG4-1, p = 0.0692; Vector vs. dgCtIP-1 + dgKU70-2, p = 0.0245; Vector vs. dgCtIP-1 + dgKU80-1, p = 0.0063; Vector vs. dgLIG4-1 + dgKU70-2, p = 0.0337; Vector vs. dgLIG4-1 + dgKU80-1, p = 0.0606; Vector vs. dgKU70-2 + dgKU80-1, p = 0.0299), the representative flow cytometry figures are shown in Fig. S3c. e Strategy for insertion of an EGFP reporter gene into the human AAVS1 locus using CRISPR-Cas9 in human cells. The SA-T2A-EGFP promoterless cassette was flanked by two AAVS1 homology arms, left arm (489 bp) and right arm (855 bp). SA, splice acceptor, T2A, a self-cleaving peptide, PA, a short polyA signal, primer F and primer R were designed for EGFP-positive events identification and sequencing. f Chromatogram and sequences of HDR-positive events. Partial donor sequences and adjacent genomic DNA sequence were represented. g–l HDR efficiency was determined in three different cell lines, HEK293 (Vector vs. dgCDK1-2, p = 0.0153; Vector vs. dgKU80-1, p = 0.0404; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.029), HEK293T (Vector vs. dgCDK1-2, p = 0.0008; Vector vs. dgKU80-1, p = 0.0227; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.0087) and HeLa (Vector vs. dgCDK1-2, p = 0.015; Vector vs. dgKU80-1, p = 0.0216; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.0004). CDK1 activation and/or KU80 repression significantly increased HDR efficiency. These cell lines were co-transfected with SA-T2A-EGFP donor and sgAAVS1-mCherry expression vectors 24 h after dgRNA-Com:CK and/or dgRNA-MS2:MPH transfection. At day 3, the frequency of EGFP+ cells within mCherry+ population were determined using FACS. Data are showed as the mean ± SEM from three independent experiments. Significance was calculated using the Paired t-test. *p < 0.05, **p < 0.01, ***p < 0.001
We next sought to determine if CDK1 and CtIP activation or LIG4, KU70, and KU80 inhibition could enhance HDR frequency for CRISPR-mediated precise gene editing. To quantitatively determine the HDR and NHEJ outcome, we firstly generated a traffic light reporter (TLR)30 stable expression HEK293 cell line that also expresses Cas9 (HEK293-Cas9-TLR) (Fig. 1c). The TLR includes a nonfunctional green fluorescent reporter in which codons 53–63 were disrupted (broken frame Venus, bf-Venus), driven by a CMV promoter. In addition, a self-cleaving peptide T2A and a red fluorescent reporter with a 2 bp frameshift (fs-mCherry) were cloned closely adjacent to the bf-Venus (Fig. 1c). With an sgRNA targeting the 5′ region of the bf-Venus, Cas9 induces DSBs, which can subsequently be repaired by two major DNA repair pathways, NHEJ or HDR. NHEJ caused indels shifting the coding frame of the T2A-mCherry, where ∼1/3 of the mutagenic NHEJ events generate in-frame functional mCherry that could be detected in cells (Supplementary Fig. S2b). However, if an intact EGFP HDR donor was provided during DSB repair, the bf-Venus could be corrected in a precise manner that leaves the succeeding fs-mCherry remaining out of frame (Supplementary Fig. S2c). Thus, this TRL reporter allows us to accurately quantify HDR and NHEJ events.
Using this TLR reporter, we transfected HEK293-Cas9-TLR cell line with dgRNA-Com:CK and/or dgRNA-MS2:MPH plasmids targeting CDK1, CtIP, LIG4, KU70, and KU80 to modulate the expression of these factors. 24 h later, cells were co-transfected with PCR EGFP HDR template and sgVenus-ECFP expression plasmids (Supplementary Fig. S3a). ECFP+ cells were gated by FACS after 48 h of transfection (Supplementary Fig. S3b), and the frequency of EGFP+ and mCherry+ cells were determined (Supplementary Fig. S3c). In the vector group, we observed 2.42% EGFP+ and 6.82% mCherry+ cells, which represented HDR-positive and NHEJ-positive events, respectively (Supplementary Fig. S3c). In contrast, the percentage of EGFP+ cells was dramatically increased after activating HDR-related genes by dgRNA-MS2:MPH, or repressing NHEJ-related genes by dgRNA-Com:CK, for all 15 single or combinatorial perturbations tested, with 12/15 perturbations reaching statistical significance (Fig. 1d; Supplementary Fig. S3c). Particularly, in the group of dgCDK1 - 2:MS2-MPH (dgCDK1–2) + dgKU80-1:Com-CK (dgKU80-1), 15.4% EGFP-positive cells were observed (Fig. 1d; Supplementary Fig. S3c). Of note, NHEJ has two major sub-classes of pathways, classical NHEJ (C-NHEJ) and micro-homology-mediated end-joining (MMEJ), also known as alternative end joining (Alt-EJ), which uses of 5–25 bp microhomologous sequences during broken ends alignment before the ends join, thus resulting in indels flanking the original break31,32. C-NHEJ is not error prone, whereas MMEJ is. Both pathways are distinct from HDR. Our assay measures the outcome of mutation acquisition, thus it measures non-classical NHEJ (MMEJ) but not C-NHEJ, where most of the repairs are not mutagenic thus is silent from the assay. Interestingly, KU70 and KU-80 repression by dgRNA showed that KU-silencing only increase template-dependent precise gene editing (HDR, as measured by Venus), but does not increase mutagenic repair (NHEJ/MMEJ, as measured by mCherry) (Supplementary Fig. S3c). Thus, suppression of KU70/KU80 may tip the balance of NHEJ (C-NHEJ and MMEJ) and HDR pathways, thereby increases the HDR efficiency. To confirm that the DSBs were repaired through HDR or NHEJ pathways, EGFP+/mCherry−, EGFP−/mCherry+ and EGFP−/mCherry− cells were cloned and the TLR sgRNA-targeting sites were sequenced. It was observed that in EGFP+/mCherry− clones, the bf-Venus gene was precisely repaired by the EGFP HDR donor without indels, whereas various indels were found in both EGFP−/mCherry+ and EGFP−/mCherry− clones (Supplementary Fig. S4a–c), together confirming the HDR and NHEJ events at genomic DNA (gDNA) level. Thus, with the robust TLR system, we found that modulating HDR factors, NHEJ factors, or their combinations can significantly enhance HDR efficiency, where both programming HDR/NHEJ by CRISPRa/i and Cas9-mediated gene editing were achieved simultaneously with a single Cas9 transgene.
The dgCDK1-2 + dgKU80-1 combination has the highest enhancement of HDR efficiency as revealed by TLR experiment among all tested groups/programs. To test its effect on CRISPR-mediated gene editing at an endogenous genomic locus, we measured the precise integration of an HDR donor SA-T2A-EGFP expression cassette into the first intron of the canonical AAVS1 locus upon Cas9/sgRNA-induced DSB (Fig. 1e). The SA-T2A-GFP was flanked by an AAVS1 left homology arm (489 bp) and a right homology arm (855 bp), where EGFP could only be expressed when the SA-T2A-EGFP was precisely recombined into the target site (Fig. 1e). We firstly transfected dgRNA-Com:CK and/or dgRNA-MS2:MPH constructs targeting CDK1 and KU80 genes using the HEK293-Cas9 cell line. 24 h later, these cells were co-transfected with SA-T2A-EGFP HDR donor template and an sgAAVS1-mCherry plasmid and then analyzed by FACS 48 h after transfection. Compared to the baseline 2.09% GFP+ cells in the mCherry+ population in vector group, the fraction of GFP+ cells from dgCDK1-2, dgKU80-1, and dgCDK1-2 + dgKU80-1 groups were significantly increased to 7.58%, 6.64%, and 15.3%, respectively (Fig. 1g, h). Quantitative result showed that HDR efficiency was enhanced over three-fold with single factor programming and over seven-fold with dual programming on an endogenous locus AAVS1 (Fig. 1g, h). We confirmed the results with two additional cell lines, with upto five-fold HDR enhancement in HEK293T and again five-fold in HeLa (Fig. 1i–l). We also designed another sgRNA for AAVS1 targeting using a same HDR template (Fig. 2a), the result showed that HDR also can be significantly improved (Fig. 2b, c). In addition, we tested another gene locus, ACTB, activation of CDK1 and repression of KU80 significantly enhanced HDR upto 4–5-fold (Fig. 2d-f). Results from all those cell lines and loci showed that HDR efficiency enhancement was most dramatic in the dgCDK1–2 + dgKU80-1 combination group. We amplified, cloned, and sequenced the endogenous AAVS1 locus and confirmed the precise integration of SA-T2A-EGFP into the anticipated target site (Fig. 1f, Supplementary Fig. S4d). Thus, in concordance with the exogenous TLR results, we observed an enhanced efficacy of precise gene targeting via HDR in the native mammalian genome.
a A scheme of insertion strategy at the human AAVS1 locus. A new AAVS1 targeting site was designed, sgAAVS1–2 was close to sgAAVS1-1 targeting site, but both used a same HDR donor template. b-c HDR efficiency at the different AAVS1 locus (Vector vs. dgCDK1-2, p = 0.0443; Vector vs. dgKU80-1, p = 0.0699; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.0044). d A scheme of insertion strategy at the human ACTB locus. e-f Flow cytometry showed that the HDR efficiency was significantly improved after activating CDK1 and repressing KU80 genes (Vector vs. dgCDK1-2, p = 0.1166; Vector vs. dgKU80-1, p = 0.0504; Vector vs. dgCDK1-2 + dgKU80-1, p = 0.0156). Significance was calculated using the Unpaired t-test. *p < 0.05, **p < 0.01
To further improve the programmability of this approach, we adapted it to additional conditional-expression modules and viral packaging system. To reduce potential side effects from constitutive activation of CDK1 or deficiency of KU80, we utilized a Tet-On system inducible by doxycycline (Dox) to control the expression of CRISPRa and CRISPRi effectors, MPH and CK, respectively. We constructed two vectors, TRE-MPH and TRE-CK (Fig. 3a). Both vectors contain a CMV-rtTA expression cassette, when cells are treated with Dox, the rtTA protein will specifically bind to TRE3G promoter and thereby initiate the transcription of MPH or CK downstream (Fig. 3a), which will be reversibly turned off upon Dox removal. We transfected these plasmids into HEK293-Cas9 individually and in combination, then followed by G418 selection and cell cloning to obtain TRE-MPH, TRE-CK, and TRE-MPH-CK cell lines (Fig. 3b). By qRT-PCR, we determined that CDK1 and KU80 will be significantly activated or repressed, respectively, in a select set of stable cell lines (Fig. 3c, d). We chose TRE-MPH-2 and TRE-CK-4 based on their best potency of Dox-induced CDK1 activation and KU80 repression for the subsequent endogenous HDR experiment. Three different cell lines were treated with Dox for 24 h, then the SA-T2A-EGFP HDR donor for AAVS1 locus and sgAAVS1-mCherry plasmid were co-transfected. After 48 h of transfection, EGFP+ cells in mCherry+ population were quantified by FACS, which revealed that upon Dox treatment, the percentages of EGFP+ cells significantly increased in all three groups as compared to control (Supplementary Fig. S5a), and without any side effects for Dox (Supplementary Fig. S5b), albeit interesting with a similar four-fold enhancement possibly due to the capacity of Dox-inducible gene expression. Of note, although the transcriptional levels of CDK1 activation or KU80 repression can vary between clones, the clones with significant CDK1 activation and/or KU80 repression have increased HDR efficiency. This data suggests that the CRISPRa/i DNA repair programming can be used in conjunction of inducible expression system to allow further control of HDR enhancement.
a Diagram of TRE-MPH and TRE-CK expression vectors used to activate CDK1 and repress KU80, respectively. rtTA interact with doxycycline, the complex could bind to TRE3G promoter, which then initiate the transcription of MCP-P65-HSF1 or COM-KRAB. b The workflow of establishing inducible HDR increasing system. Activation of CDK1 and/or repression of KU80 can be achieved by simply controlling the availability of doxycycline. Dox, doxycycline; Puro, puromycin. c-e HEK293-TRE-MPH, HEK293-TRE-CK, and HEK293-TRE-MPH-CK cell lines were obtained based on HEK293-Cas9 cell line by G418 selection. Several random clones were picked for each cell line. Although the transcriptional levels of CDK1 activation or KU80 repression can vary between clones, the clones with significant CDK1 activation and/or KU80 repression have increased HDR efficiency. The transcription level of CDK1 and KU80 were determined by RT-qPCR after 2 days of doxycycline treatment. f Quantitative analysis result of HDR efficiency (HEK293-TRE-MPH vs. HEK293-TRE-MPH + Dox, p = 0.001; HEK293-TRE-CK vs. HEK293-TRE-CK + Dox, p = 0.0011; HEK293-TRE-MPH-CK vs. HEK293-TRE-MPH-CK + Dox, p = 0.0032). Data was shown as the mean ± SEM from three independent experiments. Significance was calculated using the Paired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
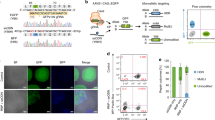
Finally, we adopt the usage of lentiviral system for stable integration of constructs for CRISPRa of DNA repair factors (Fig. 4a). We generated lentivirus-integrated cell lines expressing dgCDK1-MS2:MPH, and repeated the endogenous AAVS1 targeting experiment with introduction of HDR donor and sgAAVS1-Puro by transfection (Fig. 4b). Consistent with results above, FACS analysis again showed significant enhancement of HDR efficiency (Fig. 4c), indicating the adaptability of this DNA repair programming-mediated HDR enhancement system to viral delivery vehicles. In conclusion, our data together showed that CRISPRa/i-mediated activation and inhibition of key genes related to DNA damage repair pathways is an effective way to increase the efficiency of HDR for precise genome editing in mammalian cells. With the activation of CDK1 by dgRNA-MS2:MPH and/or repression of KU80 by dgRNA-Com:CK, the HDR efficiency can be enhanced by 4–8 fold. In this system, through combinatorial usage of sgRNA and dgRNA for different purposes, we achieved the genome editing, gene activation, and repression simultaneously simply with a single Cas9 transgene (Fig. 4d).
a The CDK1 activation plasmid was reconstructed into lentivirus backbone. Hygro, hygromycin. b HEK239FT cell was transduced with Cas9-Blast lentivirus to establish Cas9 constitutively expressed cell-line. Then the HEK239FT-Cas9 cell-line was transduced with dgCDK1-MS2:MPH lentivirus, followed by 2-3 days hygromycin selection. Finally, cells were transfected with sgAAVS1-Puro plasmid and SA-T2A-EGFP HR donor. The flow cytometry analysis was performed after 2 days' puromycin selection. Blast, blasticidin; Puro, puromycin. c The flow cytometry results demonstrated that HDR efficiency was significantly increased as compared with the vector group. d Schematic diagram representing the central idea of this study: with a single Cas9, through combinatorial usage of sgRNA and dgRNA for gene editing and CRISPRa/i on HDR/NHEJ machinery, HDR efficiency enhancement was achieved.
Discussion
Our approach provides a new way for HDR enhancement compared to other approaches. LIG4 inhibitor SCR7, which was initially defined as an anti-cancer agent33, was used to block the NHEJ pathway and improved the efficiency of Cas9-mediated HDR34,35. High-throughput screening identified two small molecule compounds (L755507 and Brefeldin A) that can enhance CRISPR-mediated HDR frequency by three-fold and two-fold, respectively36. However, these chemicals can have vast side effects or toxicity, and face availability and delivery challenges for in vivo genetic manipulations. SCR7, L755507, and Brefeldin A have insignificant effect on improvement of HDR efficiency at endogenous CTNNB1 and PRDM14 loci in human cells37. SCR7 or L755507 were shown to have minimal effects on HDR improvement of Cas9-mediated or TALEN-mediated knock-in in rabbit, HEK293 or human fibroblast cells38,39, possibly due to different modes of action or different cellular availability of these small molecules across various biological settings. Notably, severe cell death was observed after SCR7 treatment on pluripotent stem cells38. In addition, RS-1, a stimulator of human HDR protein RAD5138,39, and Ad4E1B-E4orf6, which were reported as an inhibitor of NHEJ pathway35, also been used to increase HDR efficiency and achieved a 3–6-fold effect. Interestingly, a recent study suggested that these small compounds could decrease HDR in certain settings37. Therefore, small molecular compounds are effective approaches for improving HDR efficiency, while their stability, bioavailability, side effects, toxicity, and modes of action might vary across different settings.
Another strategy to enhance the HDR efficiency is restriction of cell cycle to S and G2 phases. Timed delivery of Cas9-guide RNA ribonucleoprotein (RNP) complexes into cell cycle arrested cells using aphidicolin or nocodazole inhibitors can significantly increase HDR rates in HEK293T cells40. Similar alternative approaches have been used to boost HDR events by fusing a peptide from human Geminin with Cas9, making the fusion protein enriched during S, G2, and M phases but lower in G141,42, because cell-cycle-controlling E3 ubiquitin ligase, APC/Cdh1, is active in the late M and G1, which can ubiquitinate Geminin then leading to their degradation43,44. Cas9-hGem increased the rate of HDR up to 1.87-fold compared to wild-type Cas941. These approaches are challenging to be carried out in vivo, as cell cycles in mammalian organs are delicately controlled in a temporal and spatial manner.
Our approach is versatile and flexible, with active-Cas9-dgRNA mediating CRISPRa/i programming of DNA repair machinery, where the active Cas9 can still perform its function of generating DSB for HDR-mediated precise gene editing. These components can join force with an armamentarium of other genetic tools, such as inducible gene expression modules via simple genetic engineering. Furthermore, the CRISPRa/i constructs can be packaged into viral vectors for efficient delivery into a large repertoire of cell types. For in vivo manipulation, the construction size of CRISPRa/i is slightly larger than that traditionally used for Cas9-based HDR, and two AAV systems45 can be used for simultaneously delivering activation or/and repression components and the HDR donor template. Because DNA damage repair pathways are highly conserved, especially in mammalian species and cell types3,14. It has been demonstrated that CRISPR-mediated HDR and NHEJ occur in various cultured cell types including ES cells19, primary HSC46, and live animals in vivo47,48. Therefore, the dgRNA-based CRISPRa/CRISPRi HDR enhancement system might be worthwhile to be tested in cell types of higher clinical interest, such as ES cells and HSCs. Finally, this is a genetic approach of HDR enhancement thus can be easily adapted for in vivo settings in time-specific and tissue-specific manner, which is essential for the application of gene therapy.
Materials and methods
Generation of activation and repression plasmids
The activation plasmid dgRNA-MS2:MPH contains a U6 promoter, an MS2 gRNA scaffold, a CMV promoter, and a MCP-P65-HSF1 complex. The repression plasmid dgRNA-Com:CK consists of a U6 promoter, a Com gRNA scaffold, a CMV promoter, and a COM-KRAB complex. All key DNA fragments in these plasmids were synthesized by GENEWIZ or IDT, then cloned into pUC57, or lentiviral plasmids using general molecular cloning and Gibson assembly (NEB). dgRNAs (14-nt or 15-nt) (Supplementary Table S1) were designed to target the first 200 bp upstream of each TSS27. Five dgRNAs were designed to target each gene. TRE-MPH and TRE-CK were constructed based on dgRNA-MS2:MPH and dgRNA-Com:CK by inserting CMV-rtTA cassette and replacing CMV promoter, which drive MPH or CK expression with a TRE3G inducible promoter. For establishment of TRE-MPH, TRE-CK, and TRE-MPH-CK cell lines, we firstly transduced HEK293 cells with Cas9-expressing lentivirus to establish a constitutive Cas9 expression cell line, then transfected with TRE-MPH and/or TRE-CK plasmids followed by G418 selection and PCR identification.
Traffic light reporter (TLR) plasmid construction
TLR construct was assembled with a nonfunctional EGFP variant (bf-Venus), where codons 53–63 were disrupted, a T2A peptide, and a red fluorescent gene that has a 2-bp shifted reading frame (fs-mCherry)30. The expression cassette of Venus-T2A-mCherry was cloned in between the CMV promoter and SV40 poly (A) signal. The CRISPR-targeting site was designed at bf-Venus disrupted region. As Cas9 specifically induced DSBs, if DSBs were repaired by NHEJ pathway, ∼1/3 of repaired events generate in-frame functional mCherry. Alternatively, if DSBs were repaired by the EGFP HDR donor to generate intact Venus, the disrupted region of bf-Venus would be corrected that leaves fs-mCherry remaining out of frame.
Cell culture and transient transfection
HEK293, HEK293T, HEK293FT, and HeLa cell lines were used in this study. Cells were maintained in complete media (DMEM (Invitrogen/Thermofisher) with 10% FBS (Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL) (Life Technologies/Thermofisher)) in 37 °C, 5% CO2 incubators. Before performing the activation and repression experiments, we generated Cas9-stable expressed cell lines, HEK293-Cas9, HEK293T-Cas9, HEK293FT-Cas9, and HeLa-Cas9, either by stable integration or by transduction with Cas9 lentivirus (Cas9-Puro or Cas9-Blast), followed by puromycin or blasticidin selection. All the following activation and repression experiments were based on Cas9 stable-expression cell lines. The cells were cultured in 24-well plates (Corning) in complete media and transfected with plasmids using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s instructions. In brief, 100,000 cells/well were seeded into 24-well plates 12 h before transfection. 600 ng of plasmid encoding dgRNA-MS2:MPH or dgRNA-Com:CK were transfected with 1 μL Lipofectamine 3000 and 1 μL P3000 reagent in Opti-MEM (Invitrogen). Cells were trypsinized and re-seeded into another 24-well plate after 24 h of transfection. After 12 h of plating, cells were transfected with a 1:1 mass ratio of sgRNA plasmid and PCR HR donor. 600 ng total plasmid per well was transfected with 1 μL Lipofectamine 3000 and 1 μL P3000 reagent. Puromycin (0.5 μg/mL), Zeocin (200 μg/mL), or Blasticidin (5 μg/mL) were added after 24 h of transfection. Media was changed per 24 h with fresh pre-warmed selection media. For Tet-On induction of gene expression, cells were treated 2 days with Dox at 1 μg/mL.
Lentivirus production and transduction
Briefly, HEK293FT cells (ThermoFisher) were cultured in DMEM (Invitrogen) + 10% FBS (Sigma) media and seeded in 15-cm dishes before transfection. When cell confluent reached 80–90%, the media would be replaced by 13 mL pre-warmed OptiMEM (Invitrogen). For transfection of each dish, 20 μg transfer plasmids, 15 μg psPAX2 (Addgene 12260), 10 μg pMD 2.G (Addgene 12259), and 130 μL PEI were added into 434 μL OptiMEM, briefly vortexed, and incubated at room temperature for 10 min before being added to the 13 mL OptiMEM. The 13 mL OptiMEM would be replaced with pre-warmed 10% FBS in DMEM. Lentivirus supernatant was harvested 48 h after media change and aliquoted, and stored at −80 °C in a freezer. For Cas9-Puro or Cas9-Blast transduction, HEK293, HEK293T, HEK293FT, and HeLa cell lines were transduced with Cas9-Puro or Cas9-Blast lentivirus and supplemented with 2 μL of 2 mg/mL polybrene (Millipore) in six-well plates. The puromycin (0.5 μg/mL) or blasticidin (5 μg/mL) selection was performed for 7 days after lentivirus transduction. For dgCDK1-MS2-MPH lentivirus transduction of HEK293FT-Cas9 cell line, hygromycin (200 μg/mL) selection was performed for 2–3 days.
RT-qPCR
Cells were collected and lysed using TRIzol (Invitrogen) after 48 h of drugs treatment. Total RNA was isolated using the RNAiso Plus (Takara). cDNA synthesis was performed using the Advantage RT-for-PCR kit (Takara). RNA level was quantified by qPCR using SYBR Fast qPCR Mix (Takara) in 20 μL reaction, qPCR was carried out using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). We use the melt curve to confirm the specificity of primers. mRNA relative expression level was normalized to GAPDH expression by the ΔΔCt method.
Confocal fluorescence imaging
Before performing confocal fluorescence imaging, transfected cells were trypsinized and re-seeded on glass cover slips overnight. After aspirating the medium, cells were treated with 4% formaldehyde/PBS for 15 min for fixation, where their nuclei were stained with DAPI (CST) in PBS. EGFP or mCherry fluorescence was visualized by a confocal microscope (Zeiss LSM 800). Confocal data were analyzed by Image J software (NIH, Bethesda, MD, USA).
Flow cytometry analysis
Flow cytometric (or FACS) assays were used to evaluate the percentage of EGFP-positive or mCherry-positive cells. Briefly, HEK293-Cas9, HEK293T-Cas9, HEK293FT-Cas9, and HeLa-Cas9 cells were transfected with sgRNA plasmid and HR donor, then cultured for 72 h. The cells were digested by Trypsin without EDTA, followed by brief centrifugation and resuspension in PBS, then determining the cell density and diluting to 1 × 106 cell/mL. Finally, these samples were analyzed using a BD Fortessa or BD FACSAria flow cytometer within one hour.
Genomic DNA isolation and DNA sequencing
The transfected cells were lysed and gDNA was extracted using the DNeasy Tissue Kit (Qiagen) following the manufacturer’s instruction. For HDR-positive events identification, PCR was performed using PrimeSTAR HS DNA Polymerase (Takara) with sequence-specific primers (Supplementary Table S3) using the condition: 95 °C for 4 min; 35 cycles of 95 °C for 20 s, 60 °C for 30 s, 72 °C for 1 min; 72 °C for 2 min for the final extension. PCR products were run on 1.5% agarose gel (Biowest). The specific DNA bands were recovered using AxyPrep DNA Gel Extraction Kit (Axygen). Purified PCR products were cloned into the pMD-19 T vector (Takara) according to the standard manufacturer’s instructions or directly sequenced by specific primers. Plasmid mini-preperations were performed using the AxyPrep Plasmid Miniprep Kit (Axygen), and midi-preparations were performed using QIAGEN Plasmid Plus Midi Kit (Qiagen). All sequencing confirmations were carried out using Sanger sequencing.
Cell cycle analysis
Cells were harvested after CRISPR/dgRNAs activation or/and repression for 72 h, and prepared single cell suspension in PBS with 0.1% BSA. Washed and spinned cells at 400×g for 5 min then being resuspended with precooled 70% ethanol, fixed cells at 4 °C overnight. Washed cells in PBS, spinned cells at 500×g for 5 min then being resuspended in 500 μL PBS containing 50 μg/mL Propidium Iodide (PI), 100 μg/mL RNase, and 0.2% Triton X-100, incubation at 4 °C for 30 min. Before flow cytometry analysis, cells were passed through 40 μm cell strainer to remove cell aggregates.
CCK-8 assay
Cell viability was measured using a Cell Counting Kit-8 (CCK-8) assay (Dojindo; CK04). The transfected cells (24 h after transfection) were seeded in a 96-well plate at a density of 2.5–5 × 103 cells. Cells were incubated for 1 h with 110 μL complete DMEM media with 10 μL CCK-8 reagent for 24 h. Cell viability detection was performed by measuring the optical absorbance at 450 nm by using a multimode reader (Beckman Coulter; DTX880).
Reagent, code, and data availability
All relevant plasmids and sequencing data will be publicly deposited upon publication. No custom codes were used in this study.
Blinding statement
Investigators were not blinded for data collection or analysis. Most experiments were repeated at least three times to ensure reproducibility.
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Jasin, M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21, 8981–8993 (2002).
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004).
Karran, P. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 10, 144–150 (2000).
Haber, J. E. Partners and pathways—repairing a double-strand break. Trends Genet. 16, 259–264 (2000).
Kass, E. M. & Jasin, M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 584, 3703–3708 (2010).
Downs, J. A. & Jackson, S. P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5, 367–378 (2004).
Shrivastav, M., De Haro, L. P. & Nickoloff, J. A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134–147 (2008).
Liang, F., Han, M. G., Romanienko, P. J. & Jasin, M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95, 5172–5177 (1998).
Takata, M. et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508 (1998).
Essers, J. et al. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19, 1703–1710 (2000).
Kadyk, L. C. & Hartwell, L. H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 (1992).
Guirouilh-Barbat, J. et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14, 611–623 (2004).
Haber, J. E. In-vivo biochemistry—physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17, 609–620 (1995).
Boulton, S. J. & Jackson, S. P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996).
Lieber, M. R., Ma, Y. M., Pannicke, U. & Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4, 712–720 (2003).
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Wang, H. Y. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Heyer, W. D., Ehmsen, K. T. & Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010).
Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 12, 1051–1054 (2015).
Dahlman, J. E. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 33, 1159–U1147 (2015).
Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–U332 (2015).
Chen, X. F. et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18, 1015–U1603 (2011).
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–U506 (2007).
Certo, M. T. et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods 8, 671–U102 (2011).
McVey, M. & Lee, S. E. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538 (2008).
Chakraborty, A. et al. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 7, 13049 (2016).
Srivastava, M. et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151, 1474–1487 (2012).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–U260 (2015).
Chu, V. T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Yu, C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015).
Zhang, J. P. et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35 (2017).
Song, J. et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548 (2016).
Pinder, J., Salsman, J. & Dellaire, G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43, 9379–9392 (2015).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Gutschner, T., Haemmerle, M., Genovese, G., Draetta, G. F. & Chin, L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 14, 1555–1566 (2016).
Howden, S. E. et al. A Cas9 variant for efficient generation of indel-free knockin or gene-corrected human pluripotent stem cells. Stem Cell Rep. 7, 508–517 (2016).
Pauklin, S. & Vallier, L. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–U286 (2015).
Dever, D. P. et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Acknowledgements
This work was supported by the State Key Program of National Natural Science of China (Grant 81730101 to F.H.), The National Key Research and Development Program of China (2016YFE0125400 to F.H.), The National Natural Science Foundation of China (Grant 81603217 to L.H., Grant 81703486 to L.Y.), and NIH/NCI Center for Cancer Systems Biology (1U54CA209992 to S. C.).
Author information
Authors and Affiliations
Contributions
L.Y. conceived and initiated the study. L.Y. and C.W. designed study. C.W. and L.Y. led the major experiments and data analysis. L.H. assisted initial project discussion and cellular experiment. N.S. and D.C. assisted molecular and cellular experiments. S.C. and F.H. secure funding and jointly supervised this work. L.Y., S.C., C.W., and F.H. wrote the paper with inputs from all authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ye, L., Wang, C., Hong, L. et al. Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov 4, 46 (2018). https://doi.org/10.1038/s41421-018-0049-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41421-018-0049-7
This article is cited by
-
Development of Cas13a-based therapy for cancer treatment
Molecular Biology Reports (2024)
-
tgCRISPRi: efficient gene knock-down using truncated gRNAs and catalytically active Cas9
Nature Communications (2023)
-
Inducible expression of large gRNA arrays for multiplexed CRISPRai applications
Nature Communications (2022)
-
Screening and Analysis of Potential Genes for DNA Damage Repair and Apoptotic Signal in iPSCs Based on CRISPR–Cas9 System
Iranian Journal of Science and Technology, Transactions A: Science (2022)
-
Futuristic CRISPR-based biosensing in the cloud and internet of things era: an overview
Multimedia Tools and Applications (2022)