Abstract
Metastasis is a well-known poor prognostic factor in cancer. However, the mechanisms how long non-coding RNAs (lncRNAs) regulate metastasis in colorectal cancer (CRC) remain largely unknown. Besides, tumor-associated macrophages (TAMs) play an important role in tumor progression, yet the contribution of lncRNA-mediated crosstalk between TAMs and CRC cells to tumor progression is not well understood. In this study, we report that lncRNA RPPH1 was significantly upregulated in CRC tissues, and the RPPH1 overexpression was associated with advanced TNM stages and poor prognosis. RPPH1 was found to promote CRC metastasis in vitro and in vivo. Mechanistically, RPPH1 induced epithelial–mesenchymal transition (EMT) of CRC cells via interacting with β-III tubulin (TUBB3) to prevent its ubiquitination. Furthermore, CRC cell-derived exosomes transported RPPH1 into macrophages which mediate macrophage M2 polarization, thereby in turn promoting metastasis and proliferation of CRC cells. In addition, exosomal RPPH1 levels in blood plasma turned out to be higher in treatment-naive CRC patients but lower after tumor resection. Compared to CEA and CA199, exosomal RPPH1 in CRC plasma displayed a better diagnostic value (AUC = 0.86). Collectively, RPPH1 serves as a potential therapeutic and diagnostic target in CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most prevalent cancers in the world1. It accounted for 8% of cancer incidence and 8% of the cancer-related deaths in 20182. The poor prognosis is largely due to the distant metastasis, which accounts for about 90% of cancer-related deaths3,4. Surgical resection with or without chemo-/radiotherapy is the standard treatment of the primary tumor, but therapeutic options for distant metastasis are limited. Metastasis is a complex process, and the molecular mechanisms remain largely unknown. Therefore, it is urgently necessary to explore the molecular mechanisms and promote the development of therapy targeting metastasis.
Long non-coding RNAs (lncRNAs) are a large class of transcripts longer than 200 bp with a limited protein-coding potential5. Many studies have shown that lncRNAs participate in a series of biological processes, including epigenetic, transcriptional, and post-transcriptional levels, as well as the initiation and progression of cancers6,7,8,9. Emerging evidence suggests that lncRNAs participate in every stage of metastasis from cell migration to distant-organ colonization10. For example, colon cancer associated transcript 2 (CCAT2) is upregulated in CRC and can promote cancer cell migration and metastasis11. LncRNA-activated by TGF-β (lncRNA-ATB) can bind to the miR-200 family and IL-11 mRNA to promote the invasion-metastasis cascade12. Although many lncRNAs have been reported to be involved in tumor biological processes, the mechanisms linking metastasis and lncRNAs are still largely unknown.
Exosomes are microvesicles ranging from 70 to 120 nm in diameter and are derived from multivesicular bodies13. Exosomes participate in cells communication by transferring proteins and nucleic acids14,15. In recent studies, many exosomal proteins, miRNAs, and lncRNAs were reported to promote tumor progression16,17,18,19. For example, lncARSR can be transported by exosomes from sunitinib-resistant cells to sensitive cells, thereby promoting sunitinib resistance in renal cell carcinoma18. However, the relationships between exosomal lncRNAs and metastasis in CRC need to be further studied.
Tumor-associated macrophages (TAMs) are the most abundant cells in tumor microenvironment20,21. Numerous studies show that TAMs are associated with poor prognosis and may promote tumor progression and metastasis22,23. TAMs may become a therapeutic target in the future. But why macrophages polarize to TAMs in tumor microenvironment is puzzling.
In the present study, we found that RPPH1 is upregulated in CRC specimens and associated with advanced TNM stage and poor prognosis. RPPH1 was found to promote CRC cells migration and invasion in vitro and in vivo. Mechanistically, RPPH1 binds to β-III tubulin (TUBB3) to prevent its ubiquitination and then induces epithelial–mesenchymal transition (EMT) to promote metastasis. Furthermore, we found that RPPH1 can be transferred by exosomes to macrophages to mediate macrophage M2 polarization. Exosomal RPPH1 turned out to be upregulated in the blood plasma of CRC patients, but downregulated afterwards. These findings indicate that RPPH1 may be a potential diagnostic marker and therapeutic target in CRC.
Results
Upregulated RPPH1 is associated with poor prognosis in CRC patients
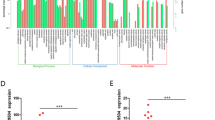
To identify lncRNAs which contribute to metastasis, next-generation sequencing (NGS) was performed on paired CRC samples and normal adjacent tissues (NATs) from seven CRC patients with liver metastasis. These seven patients’ detailed characteristics are shown in Supplementary Table 1. The flow chart of screening aimed at finding the target lncRNAs is presented in Fig. 1a. There were 212 upregulated and 447 downregulated lncRNAs with fold change > 2 or <0.5 and p < 0.05 (Fig. 1b). To identify the target lncRNAs, 31 lncRNAs were reserved with more rigorous criteria, fold change > 4, and transcript abundance >100 (Fig. 1c). Then, we chose the lncRNA with highest transcript abundance, lncRNA RPPH1, as the final target. RPPH1, Ribonuclease P RNA component H1, is the RNA component of the RNase P ribonucleoprotein24. Recent studies revealed that RPPH1 is upregulated in gastric cancer and breast cancer, but the mechanisms are not yet clear25,26. We next quantified RPPH1 in 61 paired CRC samples and NATs by quantitative real-time PCR (qRT-PCR) analysis (p < 0.001, Fig. 1d). RPPH1 expression was found to be upregulated in 78.7% (48/61) of CRC patients (Fig. 1e). Statistical analysis revealed that RPPH1 levels were strongly associated with advanced TNM stages (III and IV; p < 0.001, Fig. 1f). RPPH1 locates in chromosome 14q11.2 and the 5′ and 3′ rapid amplification of cDNA ends (RACE) assays were performed to characterize the full-length RPPH1 in CRC cells (Supplementary Fig. 1a–c). Then the coding potential of RPPH1 was analyzed by the Coding Potential Assessment Tool (CPAT), the Coding Potential Calculator (CPC), and PhyloCSF codon substitution frequency analysis (Supplementary Fig. 1d–f). All these analyses showed that RPPH1 is a non-coding RNA.
a The schematic of screening target lncRNAs. b Heatmap of the screening lncRNAs with a fold change > 2 or <0.5 and p < 0.05 from seven CRC tissues and paired normal adjacent tissues (NATs). c Heatmap of 31 lncRNAs with a fold change > 4 and transcript abundance >100. d RPPH1 RNA levels were quantified in 61 paired of CRC tissues and NATs using qRT-PCR. e Fold change of RPPH1 expression levels in 61 paired CRC tissues. f The relationship between RPPH1 expression levels and TNM stage. g Representative images of the ISH of RPPH1 expression (brown) in the paraffin-embedded CRC tissues of different TNM stages. Scale bar = 100 μm. h, i Kaplan–Meier curves for OS (h) and DFS (i) of CRC patients with low vs. high expression of RPPH1. Values are represented as mean ± SD
Furthermore, we analyzed RPPH1 expression in a large cohort of CRC patients by in situ hybridization (ISH; n = 293, Supplementary Table 2). The ISH analysis also suggested that RPPH1 expression was higher in advanced TNM stages (Fig. 1g) and was associated with metastasis (p < 0.001, Supplementary Table 2). An optimized cutoff of RPPH1 expression was generated using R (maxstat package) for the following analysis. Survival analysis showed that high RPPH1 expression was associated with poor overall survival (OS; p < 0.001) and disease-free survival (DFS) in CRC patients (p = 0.001; Fig. 1h, i). Taken together, these data suggested that RPPH1 plays an important role in CRC progression.
RPPH1 promotes CRC cell migration, invasion, and EMT in vitro
To investigate the functions of RPPH1 in CRC, we first analyzed the relative expression levels of RPPH1 in CRC cell lines (Supplementary Fig. 2a) and HCT8 and SW620 cells were chosen to perform the following assays because of their moderate RPPH1 expression levels compared to other CRC cell lines. Then we performed fluorescence in situ hybridization (FISH) to identify the location of RPPH1. As shown in Fig. 2a and Supplementary Fig. 2b, RPPH1 mainly localized in the cytoplasm. We constructed stable RPPH1 overexpression and knockdown cells in SW620 and HCT8 cells and quantified the RPPH1 levels by qRT-PCR (Supplementary Fig. 2c, d). RPPH1 overexpression obviously enhanced the migration and invasion abilities of HCT8 (Fig. 2b) and SW620 cells (Supplementary Fig. 2e). On the contrary, the RPPH1 knockdown decreased the migration and invasion abilities of the cells as assessed by migration and invasion assays (Fig. 2c, Supplementary Fig. 2f). However, the changes of RPPH1 expression had no impact on cells proliferation abilities according to the MTS assay (Supplementary Fig. 2g). While we cultured RPPH1-overexpressing HCT8 cells, we accidentally found these cells acquired mesenchyma-like morphological features (Fig. 2d). Hence, we next performed western blots and immunofluorescence assay to analyze the changes of EMT markers in CRC cells. We found RPPH1 overexpression increased the expression of mesenchymal markers N-cadherin and vimentin but reduced epithelial markers E-cadherin in SW620 and HCT8 cells (Fig. 2e–h). Likewise, the RPPH1 knockdown cells upregulated E-cadherin, but downregulated N-cadherin and vimentin (Fig. 2e–h). Furthermore, we analyzed the mRNA levels of EMT markers in CRC tissues to confirm the correlation between RPPH1 and EMT. As shown in Fig. 2i, j, we found that the RPPH1 transcript level was correlated with the vimentin mRNA level, but negatively correlated with the E-cadherin mRNA level. Altogether, those results suggested that RPPH1 may play a vital role in CRC cells metastasis.
a FISH analysis of the subcellular distribution of RPPH1 in HT29 cells. Scale bar = 20 μm. b, c Transwell migration and invasion assays in stable RPPH1 overexpression (b) or knockdown (c) HCT8 cells. Scale bar = 100 μm. d Representative images of HCT8 cells and RPPH1-overexpressing HCT8 cells. Scale bar = 40 μm. e, f The EMT effect was validated by WB analysis of epithelial or mesenchymal markers in SW620 (e) and HCT8 cells (f). g, h The EMT effect was further confirmed by immunofluorescence analysis in SW620 (g) and HCT8 cells (h). Scale bar = 40 μm. i, j The correlation between RPPH1 expression level and E-cadherin (i) or Vimentin (j) mRNA level was measured in 61 paired CRC tissues. The ∆Ct values (normalized to GUSB) were subjected to Pearson correlation analysis. Values are represented as mean ± SD. **p < 0.01 and ***p < 0.001
RPPH1 promotes CRC cell metastasis in vivo
It is well known that EMT is a crucial step in the early stages of tumor cells metastasis27 and we found that RPPH1 can promote EMT in CRC cells. Therefore, we further investigated whether RPPH1 can promote CRC cells metastasis in vivo. The SW620 cells with stable overexpression of RPPH1 or hairpin RNA (shRNA) targeting RPPH1 were injected into nude mice via tail vein (n = 6 per group). Eight weeks later, all the mice were euthanized, and we analyzed the lung metastasis. We found the lungs of the RPPH1 overexpression group were heavier than those in the control group, and the lungs in the RPPH1 knockdown group were lighter (Fig. 3a). Autopsy and hematoxylin and eosin staining (H&E) analysis also showed RPPH1 overexpression led to more metastasis in the lungs (Fig. 3b–d). Quantification of human HPRT mRNA levels in mouse lungs also suggested that RPPH1 promoted lung metastasis of CRC cells (Fig. 3e). Moreover, Kaplan–Meier analysis revealed that RPPH1 overexpression reduced the survival of mice, whereas the RPPH1 knockdown had the opposite effect (Fig. 3e). Collectively, these data meant that RPPH1 enhanced the metastasis ability of CRC.
a Wet lung weight in nude mice with SW620 cells injected via tail vein. b Autopsy of the lungs in the nude mice. c H&E of the lungs in the nude mice. Scale bar = 100 μm. d Number of metastatic nodules in the lungs from the nude mice via H&E (five sections evaluated per lung). e Expression of human HPRT1 mRNA levels relative to mouse β-actin in the lungs. f Kaplan–Meier survival curve for the nude mice. n = 6 per group. Values are represented as mean ± SD. **p < 0.01 and ***p < 0.001
RPPH1 physically interacts with TUBB3 in CRC cells
Next, we performed RNA pulldown assays to identify the protein partners of RPPH1 in order to uncover the molecular mechanism why RPPH1 could promote CRC cells metastasis. One specific band appeared on the electrophoretic gel at approximately 55 kDa in contrast to the antisense RPPH1 (Fig. 4a). Then the gel was subjected to mass spectrometry and we finally identified RPPH1-interacting protein TUBB3 according to protein mass and matched unique peptides (12/14). We then confirmed this finding by an independent immunoblot (Fig. 4b) and an RNA immunoprecipitation (RIP) assay (Fig. 4c, d). Moreover, we performed RPPH1 and TUBB3 immunofluorescence and found the co-localization of RPPH1 and TUBB3 in the cytoplasm of SW620 and HCT8 cells (Fig. 4e). After that, we constructed a series of biotin-labeled RPPH1 fragments according to its secondary structure to determine which region binds to TUBB3 (Supplementary Fig. 3a). As shown in Supplementary Fig. 3a, region 2 contains region 3. Based on the RNA pulldown as well as immunoblotting experiments, region 2 bound more TUBB3 than region 3 (Fig. 4f). Migration and invasion assays with RPPH1 fragments showed that both region 2 and region 3 could promote CRC cell migration and invasion. However, the capability to migrate through the membrane was demonstrated to be stronger in cells transfected with region 2 than those transfected with region 3 (Fig. 4g, h, Supplementary Fig. 3b, c). Moreover, the secondary structure of region 2 excluding region 3 could not be constructed in vitro, which precludes us from assessing this secondary structure’s function. Therefore, we hypothesize that the binding region of RPPH1 to TUBB3 is region 2 (111–314 nt fragment). Meanwhile, we identified that the GTPase domain of TUBB3 was responsible for the interaction with RPPH1 (Fig. 4i, Supplementary Fig. 3d). Taken together, these findings suggested that RPPH1 physically binds to TUBB3 in CRC cells.
a Biotinylated sense and antisense RPPH1 were transcribed in vitro and incubated with SW620 whole-cell lysates for RNA pulldown assays. A specific band (arrow) at about 55 kDa was excised after sliver staining. b Immunoblotting for specific associations of TUBB3 with RPPH1. c, d RNA immunoprecipitation (RIP) assays were performed using antibodies against TUBB3. qRT-PCR (c) and RT-PCR (d) assays were used to detect RPPH1. e Confocal FISH images demonstrated RPPH1 co-localization with TUBB3 in the cytoplasm in SW620 and HCT8 cells. Scale bar = 10 μm. f Serial deletions of RPPH1 were used to identify the domains of RPPH1 that bond to TUBB3 by the RNA pulldown assays. g, h Transwell migration and invasion assays in stable RPPH1 overexpression or truncated RPPH1 SW620 cells. Scale bar = 50 μm. i RIP assays were used to identify the domains of TUBB3 that bind to RPPH1 in SW620 cells. Values are represented as mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001
RPPH1 promotes CRC cell migration, invasion, and EMT via TUBB3
Previous studies have revealed that TUBB3 is overexpressed in CRC and is associated with EMT28. In our study, we found RPPH1 promotes CRC cell metastasis and physically interacts with TUBB3. Hence, we wondered whether RPPH1 performed this function via TUBB3. We identified RPPH1 levels had no effect on TUBB3 mRNA levels in CRC cells (Fig. 5a, Supplementary Fig. 4a), whereas TUBB3 protein levels increased when RPPH1 was overexpressed. In agreement with these data, the TUBB3 protein levels diminished when RPPH1 was knocked down (Fig. 5b). We next treated SW620 and HCT8 cells with the protein synthesis inhibitor cycloheximide (CHX) and measured the stability of the TUBB3 protein. The protein stability increased when RPPH1 was upregulated and decreased when RPPH1 was silenced (Fig. 5c, d, Supplementary Fig. 4b, c). In addition, we proved that proteasome inhibitor MG132 attenuates this effect (Fig. 5e). As expected, the ubiquitination of TUBB3 increased when RPPH1 was knocked down and decreased when RPPH1 was overexpressed in SW620 (Fig. 5f) and HCT8 cells (Supplementary Fig. 4d). To determine whether TUBB3 can promote CRC cells migration, invasion, and EMT, we constructed TUBB3 overexpressed and knockdown CRC cell lines (Fig. 5g). Migration and invasion assays revealed that TUBB3 overexpression enhanced CRC cells migration and invasion abilities (Fig. 5h, i, Supplementary Fig. 4e, f). Consistently with these data, expression of mesenchymal markers N-cadherin and vimentin increased whereas epithelial markers E-cadherin diminished in western blotting when TUBB3 was upregulated. The TUBB3 knockdown had the opposite effect (Fig. 5j, Supplementary Fig. 4g). After that, we investigated whether RPPH1 promotes migration, invasion, and EMT via TUBB3. When RPPH1 was overexpressed, the TUBB3 knockdown reduced N-cadherin and vimentin levels but increased E-cadherin expression. TUBB3 attenuated the N-cadherin and vimentin downregulation when RPPH1 was repressed (Fig. 5k, Supplementary Fig. 4h). Altogether, these results provide evidence that RPPH1 promotes migration, invasion, and EMT via TUBB3.
a Relative TUBB3 mRNA levels were quantified by qRT-PCR in SW620 cells with stable RPPH1 overexpression or knockdown. 18S rRNA served as the control. b WB analysis for TUBB3 protein levels in stable RPPH1 overexpression or knockdown SW620 and HCT8 cells. c, d SW620 cells with stable RPPH1 overexpression or knockdown were treated with cycloheximide (CHX, 50 μg/ml) for the indicated times and TUBB3 protein levels were analyzed via WB analysis. e Stable RPPH1 knockdown SW620 and HCT8 cells were treated with MG132 (25 μM) for 12 h and then TUBB3 protein levels were analyzed via WB analysis. f Cell lysates form stable RPPH1 overexpression or knockdown SW620 cells treated with MG132 for 12 h were immunoprecipitated (IP) with either control IgG or TUBB3 antibody and then immunoblotted for ubiquitin and TUBB3. g WB analysis for TUBB3 protein levels in stable TUBB3 overexpression or knockdown SW620 and HCT8 cells. h, i Transwell migration and invasion assays in stable TUBB3 overexpression (h) or knockdown (i) HCT8 cells. j The EMT effect was validated by WB analysis of epithelial or mesenchymal markers in SW620 cells. k Rescue assays for WB analysis of the change of EMT markers were performed in SW620 cells with RPPH1 and TUBB3 changing. Values are represented as mean ± SD. NS no significant. **p < 0.01, and ***p < 0.001
CRC cell-derived exosomal RPPH1 induces macrophages M2 polarization in vitro
Intriguingly, we found RPPH1 is abundant in blood exosomes of 12 CRC patients according to the exoRBase database (Supplementary Fig. 5a)29. Therefore, we further explored the underling mechanism of the potential link between RPPH1 with tumor environment. Before the exosomes isolation, we performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay to evaluate the cell condition and eliminate apoptotic bodies or random cell debris (Supplementary Fig. 5b). Next, we isolated the exosomes from supernatants of CRC cells and identified them via transmission electron microscopy (TEM), Particle Metrix (PMX) and western blotting through ultracentrifugation. As presented in Fig. 6a, b, the typical particles were about 100 nm in diameter. We next identified the presence of exosomal markers TSG101 and CD9 (Fig. 6c). These data indicated we successfully isolated CRC cell exosomes. We next performed RT-PCR and polyacrylamide gel electrophoresis (PAGE) to confirm the existence of RPPH1 in exosomes. When the exosomes were treated with RNase, RPPH1 levels remained unchanged. However, when the samples were treated with RNase and Triton X-100, RPPH1 levels dramatically diminished (Fig. 6d). Altogether, we proved that RPPH1 was mainly packaged in exosomes.
a, b Exosomes isolated from supernatants of CRC cells via ultracentrifugation were detected by transmission electron microscopy and Particle Metrix. Scale bar = 100 μm. c WB analysis of exosomes markers TSG101 and CD9. Exosomes extracts were treated with Triton X-100 + proteinase K or proteinase K alone. d PAGE analysis of RPPH1 levels in exosomes after RT-PCR treated with RNase (2 mg/ml) alone or combined with Triton X-100 (0.1%) for 15 min. e, f Confocal microscopy and flow cytometric analysis of the internalization of PKH26-labeled exosomes in MDMs. Scale bar = 10 μm. g Flow cytometric analysis of the expressions of CD206/HLA-DR in macrophages treated with different concentrations of exosomes isolated from supernatants of CRC cells. Numerical values denote the relative fluorescence intensity. h PAGE analysis of RPPH1 levels in exosomes isolated from supernatants of stable RPPH1 overexpression or knockdown cells. i Confocal microscopy of the macrophages treated with exosomes with different RPPH1 levels. Scale bar = 10 μm. j Flow cytometric analysis of the expressions of CD206/HLA-DR in macrophages treated with exosomes with different RPPH1 levels. Numerical values denote the relative fluorescence intensity
Exosomes play a vital role in the cell communication by transporting lncRNAs18 and TAMs are the most abundant cells in tumor microenvironment20,21. Therefore, we hypothesized that exosomal RPPH1 strongly involved in the communication between cancer cells and TAMs. First, to explore whether exosomes could be internalized into macrophages, PKH26-labeled exosomes were incubated with human monocyte-derived macrophages (MDMs) for 12 or 24 h and then confocal microscopy (Fig. 6e) and flow cytometric analysis (Fig. 6f) revealed rapid uptake of exosomes by MDMs. For functional assays, MDMs were treated with 20 or 40 μg/ml exosomes (20 ng/ml IL-4 treatment as a positive control) for the indicated times. In response to exosomes stimulation, these cells exhibited a CD206high/HLA-DRlow phenotype (Fig. 6g), with a stretched and elongated cellular morphology (Supplementary Fig. 5c). In comparation with untreated MDMs, MDMs treated with exosomes expressed significantly more M2 macrophages markers CCL17, CCL18, CXCL8, IL-10, and TGF-β, but there were no significant changes of M1 macrophage markers TNF-α, IL-6, and IL-1b via qRT-PCR (Supplementary Fig. 5d). These findings verified that CRC cell exosomes promoted macrophages M2 polarization. Then we isolated the exosomes from the supernatants of RPPH1 overexpressed or knockdown CRC cells and detected the exosomal RPPH1 levels were consistent with these changes in the parental cells (Fig. 6h). The macrophages exhibited a CD206high/HLA-DRlow phenotype, M2 morphology, and upregulation of M2 markers, when treated with exosomes with higher RPPH1 levels. As expected, treatment with exosomes with lower RPPH1 levels increased the expression of M1 markers and decreased M2 markers (Fig. 6i, j, Supplementary Fig. 5e). Overall, CRC cell-derived exosomal RPPH1 promotes macrophages M2 polarization in vitro.
Exosomal RPPH1 mediates macrophages M2 polarization to promote CRC cell metastasis and proliferation in vivo
To clarify the effect of exosomal RPPH1-mediated macrophages M2 polarization on CRC cells, tumor cells labeled with green fluorescent protein (GFP) and MDMs treated with exosomes containing different RPPH1 levels or with IL-4 were subcutaneously injected into nude mice (n = 6 per group). We measured the tumor volumes every 3 days. We found that MDMs treated with exosomes containing higher RPPH1 levels increased the volume and weight of tumors (Fig. 7a–c). Three weeks later, flow cytometric analysis was conducted to analyze circulating tumor cells (CTCs) from whole-blood samples. As expected, MDMs treated with exosomes containing higher RPPH1 levels increased the number of CTCs as compared to the control group. Conversely, MDMs treated with exosomes containing lower RPPH1 levels decreased the number of CTCs (Fig. 7d, e). To further demonstrate the effect of exosomal RPPH1-mediated macrophages M2 polarization on CRC cells, we examined the protein levels of E-cadherin, Vimentin, Ki67, and CD206 in the xenografts by immunohistochemical analysis (IHC). Similarly, MDMs treated with exosomes containing higher RPPH1 levels raised the levels of Vimentin and Ki67, but decreased the expression of E-cadherin in the xenografts (Fig. 7f). In summary, RPPH1 promoted CRC cells metastasis and proliferation via mediating macrophages M2 polarization.
a Tumors in nude mice subcutaneously injected with SW620 and different RPPH1 levels exosomes or IL-4-treated MDMs. n = 6 per group. b Tumor volumes were measured in different groups every 3 days. n = 6 per group. c Tumor weight was measured 3 weeks later when the mice were sacrificed. n = 6 per group. d Flow cytometric analysis was performed to analyze the GFP-labeled CTCs from whole blood when the mice were sacrificed. e Statistics of the flow cytometric analysis. n = 4 per group. f H&E staining and immunohistochemistry of E-cadherin, Vimentin, Ki67, and CD206. Scale bar = 50 μm. Values are represented as mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001
Circulating exosomal RPPH1 may be a diagnostic biomarker of CRC
To further explore the clinical significance of exosomal RPPH1, we isolated exosomes from CRC patients’ and healthy donors’ plasma samples. TEM and PMX analysis revealed the size and shape of vesicles from plasma was consistent with that of exosomes (Fig. 8a, b). Moreover, we performed western blots and detected exosomal markers CD63, TSG101, and CD9 (Fig. 8c). Then, we determined the exosomal RPPH1 levels in plasma samples of 52 CRC patients and 41 healthy donors by qRT-PCR. We found the exosomal RPPH1 levels were significantly higher in CRC patients than healthy donors (Fig. 8d). Besides, we measured the exosomal RPPH1 levels in 20 CRC patients 3 months after tumor resection. We found the exosomal RPPH1 levels dramatically decreased in post-operative samples in comparison with their paired pre-operative samples (Fig. 8e). Finally, we compared the diagnostic power between exosomal RPPH1 and the conventional (wildly accepted) tumor markers CEA, CA199, and CA125 via receiver operating characteristic (ROC) curve analyses. As shown in Fig. 7f, the area under ROC curve (AUC) for exosomal RPPH1, CEA, CA199, and CA125 was 0.856 (95% CI = 0.783–0.930, p < 0.001), 0.790 (95% CI = 0.698–0.882, p < 0.001), 0.544 (95% CI = 0.437–0.672, p = 0.371), and 0.654 (95% CI = 0.540–0.767, p = 0.371) respectively. These data indicated that exosomal RPPH1 was superior to the traditional tumor markers. Altogether, circulating exosomal RPPH1 is a good diagnostic biomarker for CRC.
a, b Exosomes isolated from CRC patients’ plasma samples by Exosome RNA Amplification Kit (SBI) were detected by transmission electron microscopy and Particle Metrix. Scale bar = 100 μm. c WB analysis of exosomes markers CD63, TSG101, and CD9. d Relative expression of exosomal RPPH1 in CRC patients (n = 52) and healthy donors (n = 41) via qRT-PCR. Exogenous λ polyA RNA served as the control. e Levels of exosomal RPPH1 in CRC patients’ plasma changes before surgery (pre-operation) and 30 days after tumor resection (post-operation). f ROC curves for detection of CRC using exosomal RPPH1, CA125, CEA, or CA199. g A schematic model of RPPH1 functions in tumor metastasis
Discussion
Metastasis of CRC is a major contributing of poor prognosis30. LncRNAs have been reported to participate in diverse biological processes, including metastasis10,31. In the present study, we found that lncRNA RPPH1 which is associated with advanced TNM stage and poor prognosis. Consistently, RPPH1 enhanced CRC cells migration and invasion in vitro and in vivo. Mechanistically, RPPH1 binds to TUBB3 to prevent its ubiquitination and then induces EMT in CRC cells. Furthermore, we found RPPH1 can be transmitted by exosomes to macrophage to mediate macrophage M2 polarization, thereby promoting CRC cell metastasis and proliferation. These findings illustrate that RPPH1 promotes CRC cell metastasis by functioning within cells and changing the tumor microenvironment.
TUBB3, β-III tubulin, is mainly expressed in neuronal tissues and testes32. Recent studies revealed that TUBB3 is overexpressed in solid tumors, such as breast, ovarian, testis, and colorectal cancer28,33. Furthermore, TUBB3 overexpression is a marker of taxane- or vinca-alkaloid-based drug resistance and is associated with poor outcome in various epithelial tumors, including CRC34,35,36. In this study, we explained why TUBB3 is overexpressed in CRC. RPPH1 binds to TUBB3 to enhance its stability and prevent its ubiquitination. Previous study has found TUBB3 can modulate the behavior of Snail overexpressed cells28. In our study, we also proved TUBB3 can activate the Snail pathway, thus promoting EMT. TUBB3 facilitates the transport and proper localization of N-cadherin to the plasma membrane37, but how TUBB3 activates the Snail pathway or promotes EMT remain largely unknown.
Our study offers an important finding that RPPH1 can be transported by exosomes to macrophages to promote their polarization. TAMs are the most abundant cells in tumor microenvironment20,21. Epidemiological studies suggest that TAMs density has a strong association with the poor prognosis in various tumors22,23. TAMs promote tumor growth and metastasis mostly by secreting tumor-promoting factors, such as inflammatory cytokines38,39. In this study, we established that exosomes isolated from CRC cells supernatants can be internalized by macrophages and exosomal RPPH1 exerts an important influence on this process. Moreover, we demonstrated that the levels anti-inflammatory cytokines increased and those of pro-inflammatory cytokines decreased when macrophages were treated with exosomes. The number of CTCs increased as compared to the control. Furthermore, we found TAMs can promote tumor metastasis and growth in an animal experiment. Even though we observed RPPH1 had no effect on CRC cells proliferation in vitro, RPPH1 could be transmitted by exosomes to promote macrophages M2 polarization to promote CRC cells proliferation.
Early diagnosis and treatment of CRC can improve the prognosis and OS significantly40. However, there is no effective biomarker for the early diagnosis of CRC. As is known to all, CEA is the most convincing biomarker at present. Nevertheless, CEA has low sensitivity and specificity, especially in the early stage diagnosis. Numerous studies suggest that tumor cells can secrete exosomes containing lncRNAs into body fluids, such as blood41,42. Hence, exosomal lncRNAs could be meaningful biomarkers for the diagnosis. Our study indicates that the exosomal RPPH1 level is higher in the plasma of CRC patients relative to healthy donors. Meanwhile, the RPPH1 level significantly decreased after tumor resection. In addition, we compared the plasma exosomal RPPH1 with the traditional biomarkers, such as CEA, CA199, and CA125. The AUC for exosomal RPPH1 was 0.856, which is better than the AUCs of the traditional biomarkers. This means plasma exosomal RPPH1 exhibits higher sensitivity and specificity. Moreover, we believe that plasma exosomal RPPH1 can be a new promising biomarker for the diagnosis and prognosis biomarker for CRC.
In summary, we provided the evidence that RPPH1 overexpression is associated with poor CRC prognosis. RPPH1 promotes CRC cells metastasis by binding to TUBB3, thus inhibiting its ubiquitination and enhancing exosomes-mediated macrophages M2 polarization and influences the tumor microenvironment (Fig. 7g). Moreover, plasma exosomal RPPH1 may be a diagnostic marker and a therapeutic target in CRC.
Material and methods
Cell lines and cell cultures
The human CRC cell lines HCT8, SW620, HT29, and human embryonic kidney 293T cells were purchased from American Type Culture Collection (ATCC). All of the cells were cultured at 37 °C in Dulbecco’s modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific, St Peters, MO, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, St Peters, MO, USA) in a 5% CO2 atmosphere.
Patients and samples
Sixty-one paired human CRC samples and their matched NATs were used to analyze RPPH1, E-cadherin, and Vimentin RNA levels; these tissue samples were collected from the Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China. The matched NATs were obtained at least 3 cm away from the tumor. All the patients did not receive chemotherapy or radiotherapy before surgery. All tissues upon resection were immediately frozen in liquid nitrogen and stored at a −80 °C refrigerator upon resection until further use. All the patients signed informed consent. This study was approved by the Ethics Committee of the Sixth Affiliated Hospital, Sun Yat-sen University.
RNA sequencing
Total RNA was extracted from seven paired CRC samples and NATs from CRC patients with liver metastasis. The RNA purity was analyzed on a Bioanalyzer 2200 instrument (Aligent). Then the RNA was treated with RiboMinus Eukaryote Kit (Qiagen, Valencia, CA) to remove ribosomal RNA and a cDNA library was constructed. Finally deep sequencing was performed with an Illumina HiSeq 3000 (Illumina, San Diego, CA). In the screening, when the fold change > 2 with a p value < 0.05, the lncRNA was identified as an upregulated lncRNA. On the contrary, it was identified as a downregulated lncRNA when the fold change < 0.5 with a p value < 0.05. The NGS data used in the study (GSE138202) are available in a public repository from NCBI.
Vectors construction and construction of stable cell lines
Pfu Ultra II Fusion HS DNA Polymerase (Stratagene, Agilent Technologies) was used to amplify the cDNA encoding RPPH1 or TUBB3 and then the cDNA was cloned into lentiviral expression vector pSin-EF2-Pur reformed from pSin-EF2-Sox2-Pur (Addgene, Cambridge, MA, USA). The oligonucleotides to suppress RPPH1 or TUBB3 expression were designed by RiboBio (Guangzhou, China). Then they were cloned into lentiviral expression vector pLKO.1-Pur (Addgene, Cambridge, MA, USA). The plasmids were verified by sequencing. Empty vector pSin-EF2-Pur and vector pLKO.1-Pur carrying a scrambled shRNA served as a control. 293T cells were incubated with the vectors described above, psPAX2 and pMD2G (Addgene) according to the manufacturer’s instructions. The supernatant containing infectious lentivirus was filtered through 0.22 μm PVDF filters after harvesting at 24 h post transfection and then added into the plate to infect SW620 and HCT8 cells. Infection efficiency was confirmed by qRT-PCR. The methods for transfection and lentiviral infection were described in a previous study43. The oligonucleotide sequences for vector construction are listed in Supplementary Table 5.
RNA pulldown assay
Biotin-labeled full-length RPPH1 and antisense RPPH1 were synthesized in vitro with the Transcript Aid T7 High Yield Transcription Kit (Thermo Scientific)44. Then the MEGAclearTM Kit (Thermo Scientific) was employed to recycle the sequences according to the manufacturer’s instructions. The sequences were incubated with cell lysates at room temperature for 4 h, and then the biotin-labeled RNAs with their binding protein partner were pulled down by streptavidin magnetic beads (Thermo, USA) at 4 °C overnight. The proteins were separated by electrophoresis and visualized with the Coomassie Blue Staining Kit (Beyotime, China). The different bands between sense and antisense RPPH1 were identified using mass spectrometry and retrieved in human proteomic library. The oligonucleotide sequences for the RNA pulldown are listed in Supplementary Table 6.
RNA ISH and IHC scoring
We evaluated the marker staining results according to the previous study45. The staining intensity was graded 4 stages: 0 (none), 1 (weak), 2 (moderate), and 3 (strong). The percentage of expression was graded 5 stages: 0 (<5% staining), 1 (5–25% staining), 2 (25–50% staining), 3 (50–75% staining), and 4 (>75% staining). The sum of both scores served as the final score. Two pathologists performed the scoring analyses according to the above criteria. Those two pathologists were blinded to this study.
Exosome extraction and identification
Ultracentrifugation methods were applied to isolate exosomes from the supernatants of SW620 and HCT8 cells as previous study46. In brief, the cells were cultured in a complementary medium until about 80% confluence, and then the medium was replaced the defined medium without FBS. After 2 days of culture, we harvested the supernatants and centrifuged them at 300 × g for 15 min, 2000 × g for 15 min, and 10,000 × g for 30 min. The supernatants were filtrated through a 0.22 μm PVDF filter (Millipore, USA). Then the supernatants were collected to isolate exosomes by ultracentrifugation at 120,000 × g for 70 min (Beckman Coulter) twice. Particle Metrix (PMX), transmission electron microscopy (TEM), and western blotting were used to identify the exosomes.
The plasma exosomes and exosome RNA were isolated by SeraMir™ Exosome RNA Amplification Kit (SBI) according to the manufacturer’s instructions. In brief, 500 μl of plasma was used to combine with 120 μl ExoQuick incubated at 4 °C for 30 min and then we obtained the exosomes pellets by centrifuging at 13,000 r.p.m. for 2 min. The exosomes pellet was lysed with LYSIS Buffer, and next, we purified the exosome RNA. Plasma exosomes were also identified by PMX, TEM, and western blotting. The exosome RNA levels were normalized by exogenous λ polyA RNA (Takara, China) for qPCR47.
Isolation and cultivation of monocytes/macrophages
Human peripheral blood monocytes (PBMs) were isolated from healthy volunteer donors by density-gradient centrifugation with Ficoll-Hypaque (Pharmacia, Peapack, NJ) as previously described48,49. PBMs were seeded at 2 × 106/ml in 24-well plates in DMEM medium (GIBCO) with 10% heat-inactivated human AB serum (Gemini Bio- Products, West Sacramento, CA), 50 U of penicillin per ml, 50 μg of streptomycin per mL, 2 mM l-glutamine, and 100 ng/ml human M-CSF. By repeated gentle washing with a warm medium and replacement of supernatants, nonadherent cells were removed after 5 days of culture. After that, the macrophages were treated with 20 ng/ml recombinant IL-4 (Peprotech) or CRC cells exosomes for one day. The macrophages surface maker CD206 (eBioscience), CD68 (eBioscience), and HLA-DR (eBioscience) were analyzed by flow cytometry (BD Biosciences, New Jersey, USA).
Animal experiments
The animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University, Guangzhou, China. Male BALB/c nude mice (4–5 weeks old) were purchased from the Animal Experiment Center of Sun Yat-Sen University and were randomized into control and experimental groups. Subcutaneous tumor growth assays (n = 6 per group) were performed as previously study50. Subcutaneous tumors were subjected to IHC. We analyzed the GFP-positive CTCs in PBMCs of mice by flow cytometry as the previously study12. A tail vein injection model was used for lung colonization assays (n = 6 per group). The investigator was blinded to the group allocation.
Statistics analysis
GraphPad Prism Software (GraphPad Software, La Jolla, CA, USA) was used to perform statistics analysis. Two-tailed Student’s test, ANOVA with Tukey’s multiple comparisons post-test, and Pearson’s correlation analysis were performed for statistical comparisons. All statistics analysis data are expressed as mean ± standard error of the mean. All p values were two-sided and a p value < 0.05 was considered statistically significant. All experiment was performed at least three times.
The detailed methods of the procedures are provided in Supplementary methods.
Change history
16 June 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Fidler, I. J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458 (2003).
Christofori, G. New signals from the invasive front. Nature 441, 444–450 (2006).
Martens-Uzunova, E. S. et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 65, 1140–1151 (2014).
Hacisuleyman, E. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 (2014).
Batista, P. J. & Chang, H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013).
Seiler, J. et al. The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res. 45, 5458–5469 (2017).
Liu, Q. et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 41, 4976–4987 (2013).
Huang, Q., Yan, J. & Agami, R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev. 37, 75–81 (2018).
Ling, H. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 23, 1446–1461 (2013).
Yuan, J. H. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014).
Thery, C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15 (2011).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Peinado, H. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 (2012).
Madhavan, B. et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J. Cancer 136, 2616–2627 (2015).
Pope, S. M. & Lasser, C. Toxoplasma gondii infection of fibroblasts causes the production of exosome-like vesicles containing a unique array of mRNA and miRNA transcripts compared to serum starvation. J Extracell. Vesicles 2, https://doi.org/10.3402/jev.v2i0.22484 (2013).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018).
Allavena, P., Sica, A., Solinas, G., Porta, C. & Mantovani, A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit. Rev. Oncol. Hematol. 66, 1–9 (2008).
Chittezhath, M. et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 41, 815–829 (2014).
Zhou, W. et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 17, 170–182 (2015).
De Palma, M. & Lewis, C. E. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell 23, 277–286 (2013).
Baer, M., Nilsen, T. W., Costigan, C. & Altman, S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 18, 97–103 (1990).
Xia, T. et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 4, 6088 (2014).
Zhang, Y. & Tang, L. Inhibition of breast cancer cell proliferation and tumorigenesis by long non-coding RNA RPPH1 down-regulation of miR-122 expression. Cancer Cell Int. 17, 109 (2017).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Sobierajska, K. et al. Beta-III tubulin modulates the behavior of Snail overexpressed during the epithelial-to-mesenchymal transition in colon cancer cells. Biochim. Biophys. Acta 1863, 2221–2233 (2016).
Li, S. et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 46, D106–D112 (2018).
Mina, L. A. & Sledge, G. W. Jr. Rethinking the metastatic cascade as a therapeutic target. Nat. Rev. Clin. Oncol. 8, 325–332 (2011).
Hombach, S. & Kretz, M. Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 937, 3–17 (2016).
Cleveland, D. W. & Sullivan, K. F. Molecular biology and genetics of tubulin. Annu. Rev. Biochem. 54, 331–365 (1985).
Kavallaris, M., Burkhart, C. A. & Horwitz, S. B. Antisense oligonucleotides to class III beta-tubulin sensitize drug-resistant cells to Taxol. Br. J. Cancer 80, 1020–1025 (1999).
Ferrandina, G. et al. Class III beta-tubulin overexpression is a marker of poor clinical outcome in advanced ovarian cancer patients. Clin. Cancer Res. 12, 2774–2779 (2006).
Seve, P. et al. Expression of class III {beta}-tubulin is predictive of patient outcome in patients with non-small cell lung cancer receiving vinorelbine-based chemotherapy. Clin. Cancer Res. 11, 5481–5486 (2005).
Leandro-Garcia, L. J. et al. Tumoral and tissue-specific expression of the major human beta-tubulin isotypes. Cytoskeleton (Hoboken) 67, 214–223 (2010).
Wawro, M. E. et al. Tubulin beta 3 and 4 are involved in the generation of early fibrotic stages. Cell Signal. 38, 26–38 (2017).
Ishihara, D. et al. Wiskott-Aldrich syndrome protein regulates leukocyte-dependent breast cancer metastasis. Cell Rep. 4, 429–436 (2013).
Nie, Y. et al. Tumor-associated macrophages promote malignant progression of breast phyllodes tumors by inducing myofibroblast differentiation. Cancer Res. 77, 3605–3618 (2017).
Edwards, B. K. et al. Annual report to the nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer 120, 1290–1314 (2014).
Laurent, L. C. et al. Meeting report: discussions and preliminary findings on extracellular RNA measurement methods from laboratories in the NIH Extracellular RNA Communication Consortium. J. Extracell. Vesicles 4, 26533 (2015).
Li, Z. et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 37, 3822–3838 (2018).
Yuan, R. et al. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin. Exp. Metastasis 32, 169–179 (2015).
Tsai, M. C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Ohara, Y. et al. Histological and prognostic importance of CD44(+) /CD24(+) /EpCAM(+) expression in clinical pancreatic cancer. Cancer Sci. 104, 1127–1134 (2013).
Greening, D. W., Xu, R., Ji, H., Tauro, B. J. & Simpson, R. J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 1295, 179–209 (2015).
Gilsbach, R., Kouta, M., Bonisch, H. & Bruss, M. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques 40, 173–177 (2006).
Song, E. et al. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77, 7174–7181 (2003).
Smith, M. P. et al. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 4, 1214–1229 (2014).
Yuan, J. H. et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 54, 2025–2035 (2011).
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81870383), Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (No. 2018GZR0201005), Guangdong Natural Science Foundation (No. 2017A030313785), Science and Technology Planning Project of Guangzhou City (No. 201804010014), Science and Technology Planning Project of Guangdong Province (No. 2015B020229001), and National Key R&D Program of China (No. 2017YFC1308800).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by B. Rotblat
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liang, Zx., Liu, Hs., Wang, Fw. et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis 10, 829 (2019). https://doi.org/10.1038/s41419-019-2077-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-2077-0
This article is cited by
-
Cancer-derived exosomes as novel biomarkers in metastatic gastrointestinal cancer
Molecular Cancer (2024)
-
Exosome circATP8A1 induces macrophage M2 polarization by regulating the miR-1-3p/STAT6 axis to promote gastric cancer progression
Molecular Cancer (2024)
-
Two-stage association study of mitochondrial DNA variants in allergic rhinitis
Allergy, Asthma & Clinical Immunology (2024)
-
Proteomic characterization identifies clinically relevant subgroups of soft tissue sarcoma
Nature Communications (2024)
-
lncRNA BANCR promotes the colorectal cancer metastasis through accelerating exosomes-mediated M2 macrophage polarization via regulating RhoA/ROCK signaling
Molecular and Cellular Biochemistry (2024)