Abstract
Accumulating evidence from clinical and epidemiological studies has highlighted the close correlation between the individual risk of cancer and nervous system diseases. The expression of neuronal pentraxin 2 (NPTX2) is absent in Alzheimer’s disease, anxiety, and depression. Herein, we found that NPTX2 mRNA and protein expression was significantly upregulated in colorectal carcinoma (CRC). NPTX2 expression level gradually increased with CRC progression and was closely associated with poor prognosis. In vitro and in vivo studies demonstrated that NPTX2 promoted CRC proliferation and metastasis through the activation of the Wnt/β-catenin signaling pathway. As NPTX2 receptors are absent on CRC cells, NPTX2 was shown to physically interact with frizzled class receptor 6 (FZD6) to promote β-catenin translocation into the cell nucleus, resulting in an increase in the expression of MYC, cyclin D1, snail, and N-cadherin along with a decrease in the expression of E-cadherin. Knockdown of FZD6 expression with a small-interfering RNA almost completely reversed the proliferative effects of NPTX2 on CRC development. In conclusion, NPTX2, a molecule related to nervous system diseases, promotes CRC cell proliferation and metastasis through the activation of the Wnt/β-catenin pathway via direct interaction with FZD6.
Similar content being viewed by others
Introduction
Colorectal carcinoma (CRC) is one of the most common malignant tumors of the gastrointestinal tract and ranks fourth in terms of incidence and mortality associated with human cancers1,2. In general, early-stage CRC, including stage 1 and stage 2, has a relatively good prognosis and 5-year survival rate, which could reach 70%. The prognosis of patients becomes extremely poor with the progress of CRC, especially in stage 43. The main reason for treatment failure is metastasis, which is responsible for 90% of cancer-related mortality4. The incidence of liver metastasis was most common in CRC metastasis and reached up to 50%. However, very limited clinical prevention and treatment options are available for liver metastases. Therefore, there is an urgent need to explore the mechanism underlying CRC liver metastasis to improve our understanding of the pathology as well as to design potential therapeutic strategies.
Several clinical and epidemiological studies have revealed the close correlation between individual risk of cancer and nervous system diseases5,6. While some studies have described a positive correlation, others have suggested an inverse correlation7. Therefore, further studies are warranted to address this discrepancy.
Neuronal pentraxin 2 (NPTX2) is a member of the neuronal pentraxin family and is essential for synapse formation8. Pentraxins could be divided into short pentraxin and long pentraxin based on their lengths. NPTX2 belongs to the long pentraxin group and plays a role in host immunity and acute inflammation. It is closely associated with central nervous system diseases, such as Parkinson’s and Alzheimer’s diseases9. Recent studies have found that the abnormal upregulation in NPTX2 expression was correlated with the proliferation and metastasis of clear cell renal cell carcinoma and neuroblastoma10,11. However, whether NPTX2 is involved in CRC progression and metastasis, the possible underlying mechanism remains unclear.
In the present study, we found NPTX2 expression was upregulated in CRC, which promotes colorectal cancer growth and liver metastasis by the activation of the canonical Wnt/β-catenin pathway via FZD6.
Materials and methods
Patients and samples
A total of 392 paraffin sections of CRC tissues and adjacent paired non-cancerous tissues, including 30 liver metastasis tissues, were collected to design a tissue array chip from the Department of Gastrointestinal Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University. All patients with CRC underwent surgery at the Department of Gastrointestinal Surgery, Renji Hospital, School of Medicine, Shanghai Jiao Tong University between January 2014 and January 2016. The study was approved by the Research Ethics Committee of Renji Hospital and carried out in accordance with ethical standards as formulated in the Helsinki Declaration. Informed consents were provided by all patients.
Cell culture
SW480 and LoVo (human CRC cell lines) cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in the Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin.
Small-interfering RNA (siRNA) transfection
The siRNAs for NPTX2 and frizzled class receptor 6 (FZD6) were purchased from GenePharma (Shanghai GenePharma Co., Ltd., Shanghai, China). Sequences are shown in Table S1, and experimental method was performed as previously described12.
Lentivirus transfection
Full-length human NPTX2 cDNA was transfected into CRC cell lines using a lentivirus to generate Lentivirus-NPTX2 (NPTX2-HA). Lentivirus-NC was used as the negative control (NPTX2-vector). In addition, one short-hairpin RNA (shRNA) sequence against NPTX2 was transfected into CRC cell lines to generate shRNA-NPTX2, while shNC-NPTX2 was used as the negative control. The sequences of shRNA-NPTX2 and shNC-NPTX2 were GCTCATCAACGACAAGGTTGC and TTCTCCGAACGTGTCACGT, respectively.
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Trizol was used to extract RNA, and the total RNA was reverse transcribed to cDNA by PrimeScriptTM (Takara Biomedical Technology (Beijing) Co., Ltd.). We used 18S RNA as an internal control. The sequences of the primers used are shown in Table S1. The relative expression of the target gene was calculated by the −△△Ct method.
Western blot analysis
Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer supplemented with 1% protease inhibitors and phosphatase inhibitors. Protein concentration was measured with the bicinchoninic acid (BCA) assay. Western blot analysis was performed as previously described12. NPTX2 (ab69858, Abcam), FZD6 (ab150545, ab98933, Abcam), β-catenin (ab32572, Abcam), MYC (ab39688, Abcam), cyclin D1 (CCND1; ab16663, Abcam), snail-1 (ab53519, Abcam), N-cadherin (ab76011, Abcam), E-cadherin (ab1416, Abcam), Ki-67, and proliferating cell nuclear antigen (PCNA; Proteintech Group, Inc.) primary antibodies were used. Horseradish peroxidase (HRP)-conjugated Affinipure goat anti-rabbit IgG (H + L) and HRP-conjugated Affinipure goat anti-mouse IgG (H + L) were obtained from Proteintech Group, Inc (Jackson).
Immunohistochemistry (IHC)
All tissues were paraffin-embedded and cut into 4-μm-thick sections. All sections were dewaxed with xylene and hydrated with alcohol. Sodium citrate was used for antigen retrieval, and 0.3% hydrogen peroxide (H2O2) was used to block endogenous peroxidase. After blocking non-specific sites with bovine serum albumin (BSA), all the sections were incubated with an appropriate primary antibody and secondary antibody. We used 3,3′-diaminobenzidine (DAB) kit for visualization, and hematoxylin was used to stain nuclei. All the sections were dehydrated with alcohol and sealed with neutral resin. IHC staining score was calculated based on pixel intensity; staining was scored as follows as per the staining intensity: no staining, 1; weak staining, 2; moderate staining, 3; and strong staining, 413.
Cellular immunofluorescence
A 12-well chamber was used for cellular immunofluorescence assay (IBiDi). Each well was seeded with 1.5 × 104 cells. After 1–2 days, the cells were fixed with 4% paraformaldehyde and treated with 0.2% Triton X-100 for 1 min. We used 1% bovine serum albumin (BSA) to block nonspecific binding sites. After incubation with primary and secondary antibodies, the tissue sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min and covered with a coverslip. Cellular immunofluorescence was observed through a fluorescence upright microscope and confocal microscope.
Animal model
To generate a xenograft model, 5 × 106 SW480 cells were injected into the left armpit of each null mouse. After 4 weeks, all null mice were killed and the xenograft tumors were excised. The tumor volume and weight were measured, and all tissues were fixed with 4% paraformaldehyde. To generate a liver metastasis model, all nude mice were anesthetized with 0.5% pentobarbital. The abdominal cavity was opened and 1 × 106 SW480 cells/null mouse were injected into the spleen. After 2 weeks, the mice were killed and the liver metastasis tissues were excised. All tissues were fixed with 4% paraformaldehyde.
For the generation of an orthotopic model of CRC, all nude mice were anesthetized with 0.5% pentobarbital. After opening the abdominal cavity, 1 × 106 SW480 cells/null mouse were injected into the ileocecum. After 4 weeks, the mice were killed and the tumor tissues were excised and weighed. All tissues were fixed with 4% paraformaldehyde. All animal experiments were approved by the Research Ethics Committee of Renji Hospital and adhered to the local or national requirements for the care and use of laboratory animals.
Luciferase reporter assay
Sh-NPTX2 and control CRC cells were seeded onto 96-well plates and transfected with a mixture of 100 ng of TOP (TCF reporter plasmid) reporter plasmid (Wnt/β-catenin signaling) and 10 ng of Renilla as per the protocol recommended in Lipofectamine 2000 transfection system. After 24 h, firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Promega, Madison, WI), as per the recommended protocol.
Coimmunoprecipitation (Co-IP)
Total protein was extracted from CRC cells and incubated with appropriate primary antibody overnight, followed by the addition of protein A-Sepharose beads. After extensive washing, the precipitates were subjected to western blotting for the detection of the interacting proteins. Normal rabbit IgG served as a negative control. Anti-hemagglutinin was purchased from Medical & Biological Laboratories (Nagoya, Japan). Anti-NPTX2 was obtained from Proteintech Group, Inc, while anti-FZD6 was supplied by Cell Signaling Technology, Inc.
Statistical analysis
Measurement data were presented as the mean ± standard deviation (SD). Statistical analyses were conducted by SPSS 20.0 (Chicago, IL, USA) and GraphPad Prism 5 software. Correlation of NPTX2 expression with categorical clinical variables in patients with CRC was evaluated with chi square analysis or Student’s t test. Measurement data, such as age and tumor size, were evaluated with Student’s t test, while categorical variables and ranked data, such as sex, T stage, lymph node invasion, and distant metastasis, were analyzed with chi square test. Spearman’s rank correlation was used for the analysis of two-way ordered categorical data. Survival curves were generated using the Kaplan–Meier method and analyzed by the log-rank test. Statistical significance was accepted at P < 0.05.
Results
NPTX2 is overexpressed in tumor tissues and its expression correlates with poor CRC prognosis
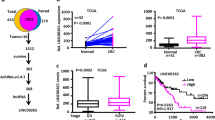
The gene expression data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets showed that NPTX2 mRNA expression was significantly increased in CRC tissues as compared with normal tissues (Fig. 1a, b). The patients with high NPTX2 mRNA expression showed an obviously shorter overall survival than those with low NPTX2 expression (Fig. 1c).
a The mRNA expression of NPTX2 in CRC and adjacent noncancerous tissues, as analyzed from TCGA dataset (Student’s t test). b The mRNA expression of NPTX2 in CRC and adjacent noncancerous tissues, as analyzed from GEO dataset (Student’s t test). c Overall survival analysis based on the mRNA expression of NPTX2 from TCGA dataset (Kaplan–Meier method and log-rank test). d, e The protein expression of NPTX2 in normal, CRC, and liver metastasis tissues in tissue microarray (392 cases of normal tissues, 392 matched cases of CRC tissues, and 30 cases of liver metastasis) (Spearman’s rank correlation). f, g NPTX2 protein expression at different pathological stages in CRC tissues from tissue microarray (34 cases of stage I, 150 cases of stage II, 127 cases of stage III, and 81 cases of stage IV CRC) (Spearman’s rank correlation). h Overall survival analysis of the protein expression of NPTX2 based on the prognostic information of patients with CRC from tissue microarray data (Kaplan–Meier method and log-rank test). Measurement data were presented as the mean ± SD. *P < 0.05; *** P < 0.0001
To evaluate the protein level of NPTX2 in CRC tissues, a tissue microarray with 392 cases of cancer and corresponding adjacent non-cancerous tissues was generated to detect the expression level of NPTX2 protein. The results of the microarray analysis showed that the expression of NPTX2 protein was significantly increased in CRC tissues as compared with adjacent noncancerous tissues. NPTX2 protein expression was obviously higher in liver metastasis than in the corresponding primary CRC (Fig. 1d, e). Further analysis showed that NPTX2 protein expression was gradually upregulated with an increase in T stages (Fig. S1A) and pathological stages (Fig. 1f, g). In addition, NPTX2 expression was significantly upregulated in CRC with lymph node invasion as compared with those without lymph node invasion; no difference was observed in NPTX2 protein expression between different lymph node invasion stages (Fig. S1B).
To investigate the clinical significance of NPTX2 in CRC, we analyzed NPTX2 expression with respect to various pathological parameters in 392 patients with CRC. The results showed that NPTX2 expression in CRC tissues was closely related to T stage, lymph node invasion, distant metastasis, and pathological stage (Table 1 and Table S2). Furthermore, survival analysis based on the expression level of NPTX2 revealed the significant decrease in the survival rate of patients with CRC that showed high NPTX2 expression as compared with those with low NPTX2 expression (Fig. 1h).
Knockdown of NPTX2 expression suppresses cell proliferation and migration in vitro and in vivo
To clarity the functional role of NPTX2 in CRC, we knocked down and overexpressed NPTX2 in two CRC cell lines, SW480 and LoVo (Fig. S2A–S2C). The results showed that the knockdown of NPTX2 expression significantly inhibited cell proliferation and cell migration abilities of both SW480 and LoVo cells (Fig. 2a–c and Fig. S2D, S2E). NPTX2 overexpression significantly enhanced cell proliferation and cell migration abilities of both SW480 and LoVo cells (Fig. 2d–f and Fig. S2F, S2G).
a The viability of SW480 and LoVo cells transfected with sh-NPTX2 or sh-control, as analyzed with CCK-8 assay (Student’s t test). b Colony-formation ability of SW480 and LoVo cells transfected with sh-NPTX2 or sh-control (Student’s t test). c Cell-migration ability of SW480 and LoVo cells transfected with sh-NPTX2 or sh-control (Student’s t test). d The viability of SW480 and LoVo cells transfected with vector or NPTX2-HA, as detected with CCK-8 assay (Student’s t test). e Colony-formation ability of SW480 and LoVo cells transfected with vector or NPTX2-HA (Student’s t test). f Cell-migration ability of SW480 and LoVo cells transfected with vector or NPTX2-HA (Student’s t test). All experiments were performed in triplicates. Measurement data were presented as the mean ± SD. *P < 0.05
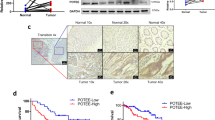
To explore the role of NPTX2 in CRC development in vivo, we applied three sets of experiments. First, sh-NPTX2 or sh-control CRC cells were subcutaneously transplanted into nude mice. As a result, the tumor volume and weight were significantly lower in the mice from sh-NPTX2 group than in those from sh-control group (Fig. 3a–c). We performed immunohistochemical staining for two markers of cell proliferation, namely, Ki-67 and PCNA, and found that the staining intensities for Ki-67 and PCNA were weaker in the tumor tissues obtained from NPTX2 knockdown cells than in those obtained from shRNA control cells (Fig. 3d). In the second set of experiment, we transplanted sh-NPTX2 or NPTX2-HA CRC cells into the cecum of nude mice to establish an orthotopic tumor model and found that the tumor weight significantly decreased after NPTX2 knockdown but obviously increased after NPTX2 overexpression (Fig. 3e). In addition, the overexpression of NPTX2 promoted ki-67 and PCNA expression, while NPTX2 knockdown inhibited ki-67 and PCNA expression in CRC (Fig. S2H, S2I). In the third experiment, we explored the role of NPTX2 in CRC metastasis; sh-NPTX2 or NPTX2-HA CRC cells were injected into the spleen of nude mice to simulate CRC metastasis to the liver through splenic vein-portal vein system. The results showed that the knockdown of NPTX2 expression inhibited CRC metastasis to the liver, while the overexpression of NPTX2 promoted CRC metastasis to the liver (Fig. 3f). Taken together, these data indicate that NPTX2 promotes CRC progression and metastasis to the liver.
a The proliferation of SW480 cells transfected with sh-NPTX2 or sh-control by subcutaneous tumor assay. b Comparison of subcutaneous tumor volume between sh-control group and sh-NPTX2 group (Student’s t test). c Comparison of subcutaneous tumor weight between sh-control group and sh-NPTX2 group (Student’s t test). d Expression of Ki-67 and PCNA in the subcutaneous tumor tissues (sh-control group [n = 5] and sh-NPTX2 group [n = 5]). e The proliferation of SW480 cells transfected with sh-NPTX2, sh-control, vector, and NPTX2-HA in an orthotopic mouse model of CRC (Student’s t test). f The proliferation of SW480 cells transfected with sh-NPTX2, sh-control, vector, and NPTX2-HA in a liver metastasis model of CRC (Student’s t test). All experiments had three replicates. Measurement data were presented as the mean ± SD. *P < 0.05
NPTX2 activates Wnt/β-catenin signaling and promotes nuclear translocation of β-catenin
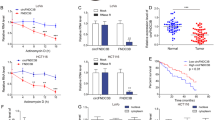
To explore the mechanism underlying the NPTX2-mediated proliferation of CRC, we examined the expression of neuronal pentraxin receptor (NPTXR), the known receptor for NPTX2, in CRC tissues and cell lines. NPTXR was rarely expressed in CRC tissues and cell lines (Fig. S3A–S3C), indicating that some other unknown receptors or interaction partners of NPTX2 may play a role in this process. To reveal the mechanism underlying NPTX2 effects in CRC development, we conducted a Gene Set Enrichment Analysis (GSEA) analysis based on NPTX2 expression in CRC as per TCGA dataset, including the mRNA sequence data from patients with CRC. The samples were divided into two groups based on the expression level of NPTX2, including high expression of NPTX2 group (NPTX2 high group) and low expression of NPTX2 group (NPTX2 high group). The result showed that the gene sets from NPTX2 high expression group were enriched in Wnt/β-catenin signaling (Fig. 4a). To confirm the correlation between NPTX2 and Wnt/β-catenin pathways, we conducted a TOP/FOP luciferase activity assay and found that the knockdown of NPTX2 expression obviously inhibited the Wnt/β-catenin pathway, while NPTX2 overexpression activated the Wnt/β-catenin pathway in CRC (Fig. 4b and Fig. S4A). However, total β-catenin protein expression showed no significant change after NPTX2 knockdown or overexpression in SW480 and LoVo cell lines (Fig. 4c and Fig. S4B–S4E). As Wnt/β-catenin activation was associated with β-catenin translocation from the cytoplasm to the nucleus, we investigated whether NPTX2 could affect β-catenin translocation. The results showed that the knockdown of NPTX2 expression significantly reduced the nuclear localization of β-catenin, while the overexpression of NPTX2 promoted nuclear β-catenin localization in both SW480 and LoVo cell lines (Fig. 4d, e and Fig. S4F). We confirmed these results in the tumor tissues from orthotopic tumor models and liver metastasis models by immunohistochemical staining for β-catenin (Fig. 4f, g). Taken together, these data indicate that NPTX2 activates Wnt/β-catenin signaling and promotes the nuclear translocation of β-catenin.
a GSEA analysis of NPTX2 expression in CRC, as evaluated from TCGA dataset. A total of 286 CRC samples were divided into two groups, including high NPTX2 expression group (143 samples) and low NPTX2 expression group (143 samples). b The fluorescence intensity of Wnt-β-catenin signaling after NPTX2 knockdown or overexpression was evaluated with a luciferase reporter assay. c Total β-catenin expression in SW480 and LoVo cells transfected with sh-NPTX2, sh-control, vector, and NPTX2-HA (Student’s t test). d Nuclear accumulation of β-catenin after NPTX2 knockdown and overexpression in SW480 and LoVo cells; e β-catenin localization in SW480 and LoVo cells transfected with sh-NPTX2 or sh-control. f β-catenin expression and localization in an orthotopic model of CRC injected with SW480 cells (sh-NPTX2 [n = 6], sh-control [n = 6], vector [n = 6], and NPTX2-HA [n = 6]). g β-catenin expression and localization in a liver metastasis model of CRC injected with SW480 cells (sh-NPTX2 [n = 6], sh-control [n = 6], vector [n = 6], and NPTX2-HA [n = 6]). *P < 0.05. All experiments were performed in triplicates. Measurement data were presented as the mean ± SD. *P < 0.05
NPTX2 physically interacts with FZD6
As mentioned above, NPTXR expression was almost absent in CRC, indicating that NPTX2 activated the Wnt/β-catenin pathway in CRC via interaction with other unknown molecules. The FZD family of proteins plays a critical role in the Wnt/β-catenin pathway and interacts with extracellular ligands, including Wnt3a and Wnt5a, to regulate the Wnt/β-catenin pathway. Therefore, we focused on FZDs and investigated whether NPTX2 interacts with FZDs in CRC. By analysis of the expression pattern of FZDs and NPTX2 in GEO dataset (GSE6988), including normal colon tissue, CRC tissue, and liver metastasis, we found only FZD6 exhibited a similar expression pattern as NPTX2, which is gradually increase from normal colon tissue, CRC tissue to liver metastasis (Fig. 5a, b and Fig. S5A). To further validate whether FZD6 could directly interact with NPTX2, we performed a co-IP assay and found that NPTX2 physically interacted with FZD6 in both SW480 and LoVo cell lines (Fig. 5c, d and Fig. S5B). In addition, immunofluorescence assay showed that NPTX2 specifically co-localized with FZD6 in CRC tissues and cell lines (Fig. 5e and Fig. S5C), thereby confirming the interaction between NPTX2 and FZD6.
a The mRNA expression of FZD6 in normal colon tissue, CRC, liver metastasis (LM) analyzed by GSE6988 dataset (Student’s t test). b The mRNA expression of NPTX2 in normal colon tissue, CRC, liver metastasis (LM) analyzed by GSE6988 dataset (Student’s t test). c, d Co-IP of NPTX2 with FZD6, a receptor of the Wnt/β-catenin pathway, in SW480 and LoVo cells; e Co-localization of NPTX2 with FZD6 in CRC tissues, as analyzed with immunofluorescence assay. All experiments had three replicates. Measurement data were presented as the mean ± SD. ***P < 0.0001
FZD6 is indispensable for the NPTX2-mediated activation of Wnt/β-catenin signaling and CRC progression
Based on the interaction between NPTX2 and FZD6, we investigated whether FZD6 serves as a key molecule in the NPTX2-mediated regulation of CRC development. The positive effects of NPTX2 on CRC proliferation and migration were almost completely abolished after the knockdown of FZD6 expression (Fig. 6a–e). Furthermore, the NPTX2-mediated nuclear localization of β-catenin also significantly reduced after the knockdown of FZD6 expression (Fig. 6f, g). The expression of some downstream target genes of Wnt/β-catenin, such as MYC, CCND1, snail-1, and N-cadherin, obviously increased after the overexpression of NPTX2, while these effects were significantly inhibited after FZD6 knockdown. E-cadherin protein expression obviously reduced after the overexpression of NPTX2, but this effect was completely abolished by FZD6 knockdown (Fig. S6A–S6E and Fig. 6h, i). Taken together, these data indicate that NPTX2 promoted CRC cell proliferation and metastasis through the activation of the Wnt/β-catenin pathway via direct interaction with FZD6.
a Viability of NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6 (Student’s t test). b Colony-formation ability of NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6. c Cell migration ability of NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6. d Colony-formation ability of NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6 (Student’s t test). e Number of migratory cells among NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6 (Student’s t test). f, g β-catenin level in the nucleus of NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6. H and I. Protein expression of β-catenin target genes in NPTX2-HA SW480 and LoVo cells transfected with si-control and si-FZD6. In all the graphs relative to experiments with cell lines, three replicates were evaluated. Measurement data were presented as the mean ± SD. *P < 0.05
Discussion
Although there is no consensus on the relationship between nervous system diseases and cancer, the overall risk of cancer was shown to decrease in the parents and siblings of the patients with nervous system diseases14. Some researchers suggest that the patients with Alzheimer’s and Parkinson’s diseases have low risk of CRC15,16. Several molecules have been found to exhibit opposite expression patterns between cancer and nervous system diseases, including TP53, adenomatous polyposis coli (APC), neuregulin-I, and phosphoinositide 3-kinase (PI3K)17. These interesting discoveries indicate that a series of tumor-related genes and nerve-related molecules show inverse expression patterns.
Another study analyzed the differential expression of genes between Alzheimer’s and non-Alzheimer’s diseases and found that the expression of C-Met, NPTX2, neurogenic differentiation factor 6 (NEUROD6), hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 (HCN1), etc., significantly decreased in Alzheimer’s disease18. Xiao et al.19 found that NPTX2 expression decreased in Alzheimer’s disease and closely correlated with cognitive functions. Li et al.20 reported the downregulation of NPTX2 expression in female mice with anxiety and depression. In the present study, NPTX2 was overexpressed in CRC tissues, and high NPTX2 expression positively associated with T stages, lymph node invasion, distant metastasis, pathological stage, and poor outcomes of patients with CRC. A study evaluating the relationship between NPTX2 expression and cancer prognosis found that NPTX2 was overexpressed in human neuroblastoma stage IV and was positively associated with poor prognosis8. Moreover, we found NPTX2 expression was higher in CRC liver metastasis than that in primary CRC tissue. Our data showed NPTX2 could promote CRC liver metastasis and enhance cell migration and proliferation abilities. These data proved that NPTX2-expressing cells had stronger migration abilities to the liver, resulting in higher expression of NPTX2 in CRC liver metastasis. Another possibility is that the metastasized CRC could secret more NPTX2 to support their colonization (growth, metastasis) by activating β-catenin signaling. It has been reported that β-catenin signaling is very important for the colonization of liver metastasis of CRC21,22.
NPTX2 as an extracellular ligand binds to its receptor, NPTXR. Extensive studies have revealed the importance of the NPTX2/NPTXR axis in nervous system diseases, involving recruitment of glutamate receptors and formation of synapses. The NPTX2/NPTXR system was shown to be upregulated in neuroblastoma and promote tumor development10. In the present study, we found that NPTX2 expression was significantly upregulated in CRC, while, to our surprise, NPTXR expression was almost completely absent in CRC. GSEA analysis revealed the close correlation between NPTX2 and Wnt/β-catenin signaling. The Wnt/β-catenin pathway is a vital regulatory pathway in CRC, and several studies have highlighted the close association between the Wnt pathway and neurite outgrowth23. Here, we demonstrate for the first time that NPTX2 promoted CRC progression and liver metastasis through the activation of Wnt/β-catenin signaling.
FZD family of proteins is the most important receptor family of the Wnt/β-catenin pathway, and abnormal expression of FZDs was closely related to carcinogenesis, especially in CRC24,25. Moreover, FZD3, FZD6, and FZD7 play vital roles in CRC tumorigenesis26,27,28. The Wnt protein family is a large family of ligands involved in the Wnt/β-catenin pathway, and Wnt3a and Wnt5a are the most important ligands in CRC. Wnt3a activates the Wnt-β-catenin pathway mainly through its interaction with FZD2/4/529. Wnt5a may regulate the Wnt-β-catenin pathway by interacting with FZD2/5/630,31. Aside from the Wnt family, others ligands were found to bind with FZDs to regulate the Wnt/β-catenin pathway32,33,34. We found that the mRNA expression of FZD6 was gradually upregulated from normal colon tissue, CRC tissue to liver metastasis, which was similar to the expression pattern of NPTX2. Subsequent, we found that NPTX2 interacted with FZD6 in CRC and demonstrated that FZD6 is indispensable for the NPTX2-mediated activation of the Wnt/β-catenin signaling and CRC progression.
There are a few limitations associated with this study. First, as the Wnt-/β-catenin pathway is a very complex system, we only revealed the interaction between NPTX2 and FZD6, but did not identify the effects of NPTX2 on other receptors or ligands, such as Wnt ligand family. Second, as an extracellular protein, NPTX2 may regulate the tumor microenvironment of CRC. For instance, the intestinal tract is rich in enteric nerve cells that may be affected by NPTX2 expression. Therefore, it would be interesting to investigate the correlation between NPTX2 expression and enteric nerve cells in CRC microenvironment.
Taken together, the present study revealed a novel molecule involved in CRC development and highlighted the inverse correlation between individual risk of cancer and nervous system diseases, owing to the differential expression of nerve-related molecules such as NPTX2.
References
Brody, H. Colorectal cancer. Nature 521, S1, https://doi.org/10.1038/521S1a (2015).
Tang, J. et al. DEAD-box helicase 27 promotes colorectal cancer growth and metastasis and predicts poor survival in CRC patients. Oncogene 37, 3006–3021 (2018).
Siegel, R. L. et al. Colorectal cancer statistics, 2017. CA: Cancer J. Clin. 67, 177–193 (2017).
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Driver, J. A. et al. Inverse association between cancer and Alzheimer’s disease: results from the Framingham Heart Study. Bmj 344, e1442 (2012).
Hedskog, L., Zhang, S. & Ankarcrona, M. Strategic role for mitochondria in Alzheimer’s disease and cancer. Antioxid. & Redox Signal. 16, 1476–1491 (2012).
Gulbinat, W. et al. Cancer incidence of schizophrenic patients. Results of record linkage studies in three countries. Br. J. Psychiatry. Supplement 18, 75–83 (1992).
Pelkey, K. A. et al. Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron 85, 1257–1272 (2015).
Moran, L. B. et al. Neuronal pentraxin II is highly upregulated in Parkinson’s disease and a novel component of Lewy bodies. Acta Neuropathol. 115, 471–478 (2008).
Bartolini, A. et al. The neuronal pentraxin-2 pathway is an unrecognized target in human neuroblastoma, which also offers prognostic value in patients. Cancer Res. 75, 4265–4271 (2015).
von Roemeling, C. A. et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 74, 4796–4810 (2014).
Xu, C. et al. SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway. Biomed. Pharmacother. 91, 1167–1177 (2017).
Yang, J. Y. et al. Decreased LKB1 predicts poor prognosis in pancreatic ductal adenocarcinoma. Sci. Rep. 5, 10575 (2015).
Ji, J. et al. Incidence of cancer in patients with schizophrenia and their first-degree relatives: a population-based study in Sweden. Schizophr. Bull. 39, 527–536 (2013).
Catala-Lopez, F. et al. Inverse and direct cancer comorbidity in people with central nervous system disorders: a meta-analysis of cancer incidence in 577,013 participants of 50 observational studies. Psychother. Psychosom. 83, 89–105 (2014).
Park, S. et al. Network comparison of inflammation in colorectal cancer and alzheimer's disease. Biomed. Res. Int. 2015, 205247 (2015).
Preti, A. & Wilson, D. R. Schizophrenia, cancer and obstetric complications in an evolutionary perspective-an empirically based hypothesis. Psychiatry. Investig. 8, 77–88 (2011).
Hokama, M. et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cereb. cortex 24, 2476–2488 (2014).
Xiao, M. F. et al. NPTX2 and cognitive dysfunction in Alzheimer’s Disease. eLife 6, https://doi.org/10.7554/eLife.23798 (2017).
Li, T. et al. Endometriosis alters brain electrophysiology, gene expression and increases pain sensitization, anxiety, and depression in female mice. Biol. Reprod. 99, 349–359 (2018).
Hugh, T. J. et al. beta-catenin expression in primary and metastatic colorectal carcinoma. Int. J. Cancer 82, 504–511 (1999).
Lee, J. H. et al. Glycoprotein 90K, downregulated in advanced colorectal cancer tissues, interacts with CD9/CD82 and suppresses the Wnt/beta-catenin signal via ISGylation of beta-catenin. Gut 59, 907–917 (2010).
Reinartz, S. et al. A transcriptome-based global map of signaling pathways in the ovarian cancer microenvironment associated with clinical outcome. Genome Biol. 17, 108 (2016).
Pez, F. et al. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J. Hepatol. 59, 1107–1117 (2013).
Kandyba, E. et al. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc. Natl. Acad. Sci. USA 110, 1351–1356 (2013).
Kim, B. K., Yoo, H. I., Kim, I., Park, J. & Kim Yoon, S. FZD6 expression is negatively regulated by miR-199a-5p in human colorectal cancer. BMB Rep. 48, 360–366 (2015).
Ueno, K. et al. Down-regulation of frizzled-7 expression decreases survival, invasion and metastatic capabilities of colon cancer cells. Br. J. Cancer 101, 1374–1381 (2009).
Dong, X. et al. RSPO2 suppresses colorectal cancer metastasis by counteracting the Wnt5a/Fzd7-driven noncanonical Wnt pathway. Cancer Lett. 402, 153–165 (2017).
Dijksterhuis, J. P. et al. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789–6798 (2015).
Gujral, T. S. et al. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 159, 844–856 (2014).
Arderiu, G., Espinosa, S., Pena, E., Aledo, R. & Badimon, L. Monocyte-secreted Wnt5a interacts with FZD5 in microvascular endothelial cells and induces angiogenesis through tissue factor signaling. J. Mol. Cell Biol. 6, 380–393 (2014).
Sun, L. et al. Sorting nexin 27 interacts with Fzd7 and mediates Wnt signalling. Biosci. Rep. 36, e00296 (2016).
Sun, Y. et al. SFRP2 augments WNT16B signaling to promote therapeutic resistance in the damaged tumor microenvironment. Oncogene 35, 4321–4334 (2016).
Lai, M. B. et al. TSPAN12 is a norrin co-receptor that amplifies Frizzled4 ligand selectivity and signaling.Cell Rep. 19, 2809–2822 (2017).
Acknowledgements
This study was supported by National Science Foundation of China (No. 81672358), Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (No. 20181708), and Outstanding academic leaders in Shanghai Health System (No. 2018BR32).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by S. Tait
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, C., Tian, G., Jiang, C. et al. NPTX2 promotes colorectal cancer growth and liver metastasis by the activation of the canonical Wnt/β-catenin pathway via FZD6. Cell Death Dis 10, 217 (2019). https://doi.org/10.1038/s41419-019-1467-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1467-7
This article is cited by
-
H3K4me3 mediates uterine leiomyoma pathogenesis via neuronal processes, synapsis components, proliferation, and Wnt/β-catenin and TGF-β pathways
Reproductive Biology and Endocrinology (2023)
-
Small extracellular vesicles promote invadopodia activity in glioblastoma cells in a therapy-dependent manner
Cellular Oncology (2023)
-
Construction of Fzd6Q152E mice through CRISPR/Cas9 technology and their reproduction and identification
Molecular Biology Reports (2022)
-
Expression and prognostic impact of FZDs in pancreatic adenocarcinoma
BMC Gastroenterology (2021)
-
Alantolactone inhibits cervical cancer progression by downregulating BMI1
Scientific Reports (2021)