Abstract
Premature ovarian insufficiency (POI) imposes great challenges on women’s fertility and lifelong health. POI is highly heterogeneous and encompasses occult, biochemical, and overt stages. MicroRNAs (miRNAs) are negative regulators of gene expression, whose roles in physiology and diseases like cancers and neurological disorders have been recognized, but little is known about the miRNAs profile and functional relevance in biochemical POI (bPOI). In this study, the expression of miRNAs and mRNAs in granulosa cells (GCs) of bPOI women was determined by two microarrays, respectively. MiR-379-5p, PARP1, and XRCC6 were differentially expressed in GCs of bPOI as revealed by microarrays. Subsequently, functional studies demonstrated that miR-379-5p overexpression inhibited granulosa cell proliferation and attenuated DNA repair efficiency. Furthermore, both PARP1 and XRCC6 showed lower levels in GCs from patients with bPOI and were identified as executives of miR-379-5p. Therefore, our data first uncovered potentially pathogenic miR-379-5p and two novel targets PARP1 and XRCC6 in bPOI, which corroborated the significance of DNA repair for POI, and brought up an epigenetic explanation for the disease.
Similar content being viewed by others
Introduction
Premature ovarian insufficiency (POI) is a hypergonadotropic disorder, which imposes great challenges on women’s fertility and lifelong health. Diagnosis is confirmed by amenorrhea for at least 4 months and elevated serum FSH levels (>25 IU/l) before 40 years of age1,2. The prevalence of POI varies by ethnicity. A latest study shows that 2.8% of the Chinese women are affected with POI3. Ovarian function decline is thought to be a continuous spectrum, three stages of POI have been described, including occult (subfertility or incipient ovarian insufficiency), biochemical insufficiency (raised concentrations of FSH, also known as transitional ovarian failure), and eventually overt (also termed as premature ovarian failure, POF [MIM 311360]) phages4,5,6. As the disorder might take several years to proceed, early detection and intervention for high-risk women may be beneficial to rescue fertility.
POI is a heterogeneous disease and two hypotheses of mechanism have long been recognized: inadequate primordial follicle pool, or accelerated depletion of oocytes and follicles7,8. Animal models suggest that dysfunction of granulosa cells is closely associated with follicle atresia, and microRNAs may play important roles during this process9,10,11,12,13. MicroRNA (miRNA) is a class of small regulatory noncoding RNA, fine-tuning gene expression post transcriptionally through mRNA degradation and inhibition of translation initiation14. It has been shown that disruption of miRNAs may contribute to physiological processes and human diseases15, likewise polymorphisms and abnormal expression of miRNAs were observed associated with POI16,17,18,19,20,21. However, given limited samples and functional evidences, miRNAs in granulosa cells (GCs) of biochemical POI (bPOI) individuals have yet to be determined.
Biochemical POI, in which stage women have regular menses but elevated FSH levels and reduced fertility, is the prior stage before follicle exhaustion completely. Here we performed miRNA and mRNA microarray in GCs from bPOI to uncover altered profiles of miRNAs and genes, then we combined bioinformatics analysis, quantitative reverse transcription (qRT)-PCR, and in vitro experiments to investigate the roles of miRNAs in bPOI. Results identified significantly upregulated miR-379-5p in GCs from bPOI patients, which suppressed cell proliferation and impaired DNA repair function through directly targeting poly ADP-ribose polymerase1 (PARP1) and X-ray repair complementing defective repair in Chinese hamster cells 6 (XRCC6), which were downregulated in GCs from the same cohort of cases.
Results
Clinical characteristics of all participants
The current study comprised 33 patients with biochemical POI and 30 age- and body mass index (BMI)-matched reproductive aged women with normal ovarian reserve. The bPOI patients undergoing in vitro fertilization/intracytoplasmic sperm injection and embryo transfer (IVF/ICSI-ET) were recruited from the Center for Reproductive Medicine, Shandong University. Inclusion criteria included (i) basal FSH (on days 2–4 of menstrual cycle) >10 mIU/ml; (ii) prior to 35 years of age; (iii) bilateral ovarian antral follicle count (AFC) <10. Thirty women with regular menstrual cycles and normal FSH level (<10 mIU/ml), who sought for infertility treatment due to tubal obstruction or male factors were enrolled as controls. Women with recurrent spontaneous abortion, chromosomal abnormality, history of chemo- or radio-therapy, ovarian surgery, or autoimmune diseases were excluded. The average age at recruitment was 30.55 ± 3.50 and 29.20 ± 3.56, respectively. Patients with bPOI exhibited typical endocrine profiles with moderately elevated bFSH levels, and dramatically less numbers of AFC and oocytes retrieved. Characteristics are summarized in Table 1.
MiR-379-5p, PARP1, and XRCC6 are differentially expressed in GCs of bPOI as revealed by microarray
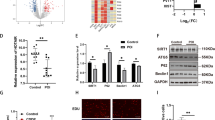
To investigate the expression profiles of miRNAs and mRNAs in GCs of bPOI patients, we first examined the change.s of miRNAs and mRNAs expression using microarrays (Exiqon miRCURY™ microarray LNA v.18.0 and Arraystar microarray v3.0) of GCs from ten bPOI and ten age- and BMI-matched normal control women. Seventy-five miRNAs (fold change (FC) >2, P < 0.05) and 3782 mRNAs (FC >2, P < 0.05; data not shown) were identified and differentially expressed between bPOI patients and control women. Of the 75 miRNAs changed in bPOI, 30 were upregulated and 45 were downregulated. Exiqon array data are available in the Gene Expression Omnibus database under accession number GSE100238. To systematically evaluate how the differentially expressed miRNAs in GCs influence gene expression and cellular activities, a miRNA–mRNA conjoint analysis was conducted and generated a bipartite miRNA/mRNA regulatory network. Then, the Gene Ontology analysis was performed to classify the putative target genes into different functional groups, which revealed an appealing biological process category of DNA repair. Hsa-miR-379-5p was one of the most upregulated miRNAs uncovered by the microarray, which exhibited a significantly negative correlation with DNA repair genes PARP1 and XRCC6 (Fig. 1a).
a Heat map of miR-379-5p, PARP1, and XRCC6 based on microarray data. Each column represents a sample; red and green indicates up- and downregulation, respectively; r correlation coefficient. b, c, d Quantitative PCR (qPCR) analysis in GCs from women with and without bPOI shows that miR-379-5p increases (b), while PARP1 and XRCC6 decrease in bPOI patients (c, d). Two-tailed Mann–Whitney U-test
MiR-379-5p is located at a highly conserved imprinted DLK1-DIO3 genomic region on 14q32.31, which shows great developmental importance and signatures in schizophrenia and metabolic disease22,23,24,25. Mature miR-379-5p harbors a conserved seed sequence. Disruption of miR-379-5p has been reported in cancers, but little is known about its roles in endocrinopathy26,27,28. Both PARP1 and XRCC6 were putative targets of miR-379-5p and downregulated in bPOI through the mRNA microarray (Fig. 1a). Numerous mutations or polymorphisms in DNA repair genes have been identified causative or associated with reproductive ageing and POI, underling the importance of DNA repair function in human fecundity29,30,31,32,33,34,35,36,37,38. Consequently, DNA repair-associated genes, PARP1 and XRCC6, were chosen as targets of miR-379-5p for further study.
To validate the microarray results, we analyzed miRNAs and mRNAs expression using qRT-PCR in an independent 43 GCs that were from bPOI (n = 23) and controls (n = 20). Consistent with microarray results, qRT-PCR showed that miR-379-5p increased (FC = 1.48, P = 0.037; Fig. 1b), while PARP1 and XRCC6 decreased significantly in the GCs from patients with bPOI (FC PARP1 = 0.72, P PARP1 < 0.01, Fig. 1c; FC XRCC6 = 0.82, P XRCC6 = 0.018, Fig. 1d).
PARP1 and XRCC6 are novel targets of miR-379-5p
PARP1 and XRCC6 are potential targets of miR-379-5p through in silico analyses. To verify the regulation of miR-379-5p on target genes, qRT-PCR and western blot were performed and indicated that PARP1 and XRCC6 mRNA and protein levels were significantly reduced by forced expression of miR-379-5p in KGN cells (Fig. 2a, b). To confirm the predicted binding sites of miR-379-5p in the 3′-UTRs of PARP1 and XRCC6, we cloned the wild type and mutated 3′-UTRs of PARP1 and XRCC6 in a luciferase reporter vector. Luciferase assay demonstrated that overexpression of miR-379-5p led to a considerable decrease in PARP1 and XRCC6 with wild-type 3′-UTRs, while luciferase activity levels of mutated 3′-UTRs unchanged (Fig. 2c, d). These results suggested that PARP1 and XRCC6 were novel targets of miR-379-5p.
a, b qRT-PCR and western blot analyses indicate that PARP1 and XRCC6 mRNA (a) and protein (b) levels are significantly reduced by forced expression of miR-379-5p in KGN cells. c, d Predicted target sites and relevant mutation sequences are shown. Luciferase assay demonstrates that overexpression of miR-379-5p leads to a decrease in PARP1 and XRCC6 with wild-type 3′-UTRs, while luciferase activity levels of mutated 3′-UTRs unchanged. Asterisk (*) indicates a significant difference. *P < 0.05, two-tailed Student's t test
Forced expression of miR-379-5p leads to proliferation inhibition and DNA repair impairment
As miR-379-5p can significantly repress PARP1 and XRCC6, which participate in DNA damage response and cell cycle control39,40, we further assess whether these processes are regulated by miR-379-5p.
For cell proliferation analysis, Cell Counting Kit-8 (CCK-8) assays and 5-ethynyl-2′-deoxyuridine (EdU) staining were performed. Proliferating cell nuclear antigen (PCNA) was also detected by western blotting. CCK-8 assay revealed that overexpression of miR-379-5p led to an inhibition of proliferation in KGN cells (Fig. 3a). And, the EdU assay showed less EdU-positive cells in miR-379-5p overexpressed group (Fig. 3b, c). PCNA level was also much lower in KGN cells overexpressing miR-379-5p than negative control (Fig. 3d).
a CCK-8 assay reveals that miR-379-5p overexpression led to suppression in cell proliferation. b, c EdU assay shows less EdU-positive cells in miR-379-5p overexpressed group. d PCNA level is much lower in KGN cells overexpressing miR-379-5p than negative control. e After exposed to MMC for 6 h, more of γH2AX is found in KGN cells overexpressing miR-379-5p. After recovery for 2 and 4 h, miR-379-5p delays the DNA damage repair. f, g The clonogenic survival of HeLa cells suffering MMC treatment and 2 h’ recovery. The clonogenic survival percent of miR-379-5p mimics group is significantly less than that of negative control. Asterisk (*) indicates a significant difference. *P < 0.05, **P < 0.01, two-tailed Student's t test
To illustrate the effect of miR-379-5p on DNA repair capacity, γH2AX, acting as a bio-marker for DNA damage, was detected to evaluate cellular response to DNA damage induced by mitomycin C (MMC). After exposed to MMC for 6 h, more γH2AX was found in KGN cells overexpressing miR-379-5p, implying higher sensitivity to MMC damage existed. When the DNA damage repair kinetics compared after recovery for 2 and 4 h, respectively, miR-379-5p delayed the DNA damage repair, which demonstrated that miR-379-5p adversely affected the progress of DNA breaks repair (Fig. 3e). HeLa cells clonogenic survival assay was conducted and showed that the clonogenic survival percentage of miR-379-5p mimics group was significantly less than that of negative control (Fig. 3f, g), indicating a harmful role of miR-379-5p in maintaining cell survival after DNA damage. Taken together, our results illustrated that forced expression of miR-379-5p led to inhibition of proliferation and impairment of DNA repair.
PARP1 and XRCC6 are functional targets of miR-379-5p
To determine whether PARP1 and XRCC6 can mediate the observed biological function of miR-379-5p, we silenced PARP1 and XRCC6 in KGN cells via specific siRNAs. Western blotting confirmed an efficient decrease in the levels of PARP1 and XRCC6 protein in KGN cells 48 h post transfection (Fig. 4a). Then, CCK-8 assay and EdU staining were performed and revealed suppressed cell proliferation in PARP1 and XRCC6 knockdown group (Fig. 4b–d). Meanwhile, PCNA level was much lower in PARP1 and XRCC6 silencing cells than negative control cells (Fig. 4e).
a An efficient decrease in PARP1 and XRCC6 protein levels is confirmed by western blot in KGN cells 48 h post transfection. b, c, d Suppressed cell proliferation in PARP1 and XRCC6 knockdown group is indicated by CCK-8 assay (b) and EdU staining (c, d). e PCNA level is much lower in PARP1 and XRCC6 silencing cells than negative control cells. f After exposed to MMC for 6 h, and recovery for 2 and 4 h, PARP1 and XRCC6 silencing delayed the DNA damage repair. g, h The percent survival relative to the control is significantly lower in PARP1 and XRCC6 depletion cells. Asterisk (*) indicates a significant difference. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student's t test
Likewise, we examined the function of PARP1 and XRCC6 in DNA repair in vitro. After exposed to MMC for 6 h, and then recovery for 2 and 4 h, silencing of PARP1 and XRCC6 delayed the DNA damage repair (Fig. 4f). Coincident with the observation of clonogenic survival assay, we identified that lack of PARP1 and XRCC6 made HeLa cells more sensitive to DNA damage (Fig. 4g, h). The above results indicated that PARP1 and XRCC6 were required for protection of cells from DNA damage and efficient DNA damage repair.
Reintroduction of PARP1 and XRCC6 abrogates the miR-379-5p-induced effects partially
To further verify the functional regulation between miR-379-5p and target genes, miR-379-5p mimic-transfected KGN cells were infected with recombinant adenovirus expressing PARP1 and XRCC6. The infection efficiency was determined by western blotting (Fig. 5a). Interestingly, the roles of miR-379-5p on cell proliferation and DNA damage repair could be abolished to some extent, confirming the regulation of miR-379-5p on the two target genes (Fig. 5b–d).
a An efficient increase in PARP1 and XRCC6 protein levels is confirmed by western blot in KGN cells 48 h post co-transfection infection. b, c PARP1 and XRCC6 overexpression could promote cell proliferation. d After exposed to MMC for 6 h, more of γH2AX is found in two groups overexpressing miR-379-5p. However, after 4 h recovery, PARP1 and XRCC6 overexpression group shows similarly less γH2AX as the NC + Ad-Null group. The results uncover that PARP1 and XRCC6 is required for efficient DNA damage repair process
Discussion
In this study, two complementary microarray screenings were performed on granulosa cells derived from patients with biochemical POI for the first time, revealed miR-379-5p was expressed at higher levels in bPOI compared to normal controls. Further, in vitro experiments showed that miR-379-5p inhibited granulosa cell proliferation and sensitized GCs to DNA damage by repressing PARP1 and XRCC6, which provide new etiologic explanation for POI.
Over the past few decades, hundreds of functional miRNAs have been identified and revealed perspectives for biological processes and diseases14,15. The relationships between miRNAs and POI have been investigated previously. Association studies evaluated that miRNA polymorphisms, such as miR-146aC>G, miR-196a2T>C, miR-499A>G, and miR-449bA>G, might be associated with high risk for POI16,17. Abnormal expression of miRNAs was observed in plasma or blood from POI patients, indicating that miRNAs serving as non-invasive diagnostic tools in clinic are promising18,19,20,21. However, uncertainties still existed owing to limited sample size and/or lacking functional evidences. Here our data significantly extended these findings by first uncovering a potentially pathogenic noncoding RNA, miR-379-5p, and two novel targets PARP1 and XRCC6 in GCs of bPOI patients. Creatively, in vitro experiments provided solid evidences to illustrate the role of miR-379-5p on granulosa cells function.
Normal function of granulosa cells is indispensable for reproduction and ovarian reserve9,41,42. Accumulated DNA damage and deficient DNA repair capacity in granulosa cells and oocytes may contribute to reproductive aging in mouse, rhesus monkey, as well as homo sapiens43,44,45. As aforementioned, perturbations in DNA repair genes, such as MCM8 (OMIM 608187; POF10), MCM9 (OMIM 610098), CSB-PGBD3 (OMIM 609413; POF11), and MSH5 (OMIM 603382; POF13), are responsible for POI in human29,30,31,32,33,34. In this study, decreased PARP1 and XRCC6 were observed in patients with bPOI. PARP1 and XRCC6 are pivotal genes involved in DNA damage repair. PARP1 binds to the ends of DNA single-strand breaks and DNA double-strand breaks (DSBs) to facilitate DNA repair process39,46,47,48. XRCC6 (also referred to as Ku70) cooperates with Ku80 to form Ku heterodimer, which initiates nonhomologous end joining pathways in DSBs repair40,49. Recently, PARP1 and XRCC6 have been proved crucial for fundamental cellular processes, metabolism, ageing, and related diseases, such as cancers, diabetes, neurodegenerative, and cardiovascular diseases39,46,47,48,49. Both Parp1−/− and Xrcc6−/− mice exhibit symptoms of accelerated ageing50,51,52,53, however, little is known concerning PARP1 and XRCC6 with ovary ageing and female fecundity. With evidence of proliferation inhibition and DNA repair attenuation in GCs, PARP1 and XRCC6 were identified as direct targets and executives of miR-379-5p. Our results further corroborated the contribution of DNA repair in the etiology of POI, and a novel epigenetic explanation was brought up.
Roles of miRNAs expressed in ovarian tissues, such as follicular fluid, corpora lutea, and cumulus–oocyte complexes, have hitherto been evaluated54,55,56. However, the progress of finding key miRNAs in GCs from POI patients has been slow due to its poor collection. In contrast with overt POI, follicles have not yet been completely depleted at bPOI stage. Therefore, we collected the GCs from individuals with bPOI undergoing IVF-ET treatment. More importantly, it has been demonstrated a remarkably rapid decline of ovarian function after menses irregularity occurred, e.g., lasting 1–2 years at average57. Therefore, it is requisite to explore the phenotypes and pathogenesis in patients still at early stage of POI.
In summary, our study demonstrated altered profiles of miRNAs and mRNAs in the GCs from patients with bPOI. Functional studies further confirmed that upregulation of miR-379-5p suppressed GCs proliferation and adversely affected DNA repair function by negatively regulating expression of PARP1 and XRCC6. This study provides new viewpoint for understanding the roles of noncoding RNA for GCs function, as well as potential etiologic mechanism for POI. Additional pathogenesis researches concentrated on miRNAs regulatory networks are warranted.
Methods
Human patient samples
The study was approved by the Institutional Review Board of Center for Reproductive Medicine, Shandong University, written informed consent was obtained from all participants. The clinical characteristics of bPOI and control women were shown in Table 1. Granulosa cells were isolated from each participant and stored at −80 ℃ until processed for RNA extraction as described previously58.
MiRNA and mRNA expression profiling assays
The miRNA and mRNA expression profiling assays of the GCs from 20 participants (10 bPOI and 10 control women) were conducted by use of Exiqon miRCURYTM LNA arrays (v18.0; contains 2043 capture probes covering all human miRNAs; Exiqon, Vedbaek, Denmark) and Arraystar Human LncRNA Microarrays (V3.0; covering a total of 26,109 protein-coding transcripts; Arraystar, Inc. Rockville, MD, USA), respectively. Microarray experiments and data analyses were performed by KangChen Bio-tech, Shanghai, China.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using the Trizol reagent (Takara Bio Inc., Dalian, China) by phenol–chloroform precipitation. MiRNAs were reverse transcribed individually by using miRNAs-specific reverse transcription primers and a mixture of dNTP (10 mM, Takara Bio Inc., Dalian, China), 5× M-MLV (Moloney Murine Leukemia Virus) buffer (10 mM, Takara Bio Inc., Dalian, China), RNase inhibitor (40 U/μl, Takara Bio Inc., Dalian, China) and M-MLV reverse transcriptase (200 U/μl, Takara Bio Inc., Dalian, China). While total RNA was reversely transcribed into cDNA using PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Dalian, China) for genes validation. The real-time polymerase chain reactions were performed using LightCycler® 480 SYBR Green I Master (La Roche Ltd, CH) and carried out by Roche LightCycle® 480 (La Roche Ltd, CH). U6 RNA and GAPDH were used as endogenous controls for qPCR of miRNAs and mRNAs, respectively. Each sample was run in triplicate. Data were analyzed according to 2−ΔΔCt method. The primers can be found in Supplementary Table 1.
Cell culture and transfection
The KGN cell line (obtained from RIKEN BioResource Center, Ibaraki, Japan), a steroidogenic human granulosa-like tumor cell line59, was cultured in DMEM/F12 (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Biological Industries (BioInd), Beit Haemek, Israel) and 1% antibiotics (HyClone, Logan, UT, USA). The human embryonic kidney (HEK) 293T cell line and HeLa (human cervix carcinoma cell line) cell line were grown in DMEM High Glucose (HyClone, Logan, UT, USA) supplemented with 10% FBS (BioInd, Beit Haemek, Israel) and 1% antibiotics (HyClone, Logan, UT, USA). All cells were cultured at 37 ℃ in a humidified atmosphere of 5% CO2 in air. MiR-379-5p mimics, specific siRNAs for PARP1/XRCC6 and negative control, were designed (siRNAs for PARP1 referred to Refs. 60 and 61) and synthesized by Genepharma Inc (Shanghai, China). MiR-379-5p mimics and siRNA-MIX were transfected at 50 nM and 100 nM, respectively, using X-tremeGENE siRNA Transfection Reagent (La Roche Ltd, CH). Sequences were provided in Supplementary Table 2.
Recombinant adenoviruses
Adenoviruses were generated and purchased from VigeneBio (Shangdong, China). For in vitro experiments, cells were infected with 2 × 107 plaque-forming units (pfu)/ml. Empty virus expressing only GFP served as control (Ad-Null).
Protein extraction and western blot
Total protein was harvested in 1× SDS loading buffer and separated by sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gel and electro-transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% milk, membranes were incubated with primary antibodies overnight at 4 ℃ and horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies for 1 h at room temperature. Immunoreactive bands were detected and analyzed with ChemiDoc MP Imaging System (BIO-RAD, Richmond, CA) and Image Lab Sofware. The following antibodies were used: anti-PARP1 (Cell Signaling Technology, 9532S, 1:1000), anti-XRCC6 (Abcam, ab3114, 1:250), anti-PCNA (Santa Cruz Biotechnology, sc-56, 1:1000), anti-γH2AX (Cell Signaling Technology, 9718s, 1:1000), anti-β-actin (Proteintech, 60008-1-Ig, 1:1000), anti-α-Tubulin (Proteintech, 66031-1-Ig, 1:1000), Peroxidase-conjugated Affinipure Goat Anti-Mouse (Proteintech, SA00001-1, 1:5000), Peroxidase-conjugated Affinipure Goat Anti-Rabbit (Proteintech, SA00001-2, 1:5000).
Luciferase reporter assay
The luciferase reporter plasmid containing wild-type 3′-UTRs of PARP1 and XRCC6 were purchased from GeneCopoeia (Rockville, MD, USA). Mutagenesis was generated by site-directed mutagenesis (Quik Change Lightning Site Directed Mutagenesis Kit; Stratagene, LaJolla, CA) with the wild-type luciferase vectors as templates. Primers used were listed in Supplementary Table 2. Wild-type and mutated vectors were cotransfected with miR-379-5p mimics or negative control into HEK-293T cells using Lipofectamine 2000 (Invitrogen). Luciferase activities of cultured supernatant were measured 48 h later using Secrete-PairTM Dual Luminescence Kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s instructions.
In vitro proliferation assays
KGN cells transfected or infected with miRNAs, siRNAs, or adenovirus for 24 h were reseeded in 96-well plates for different experiment purposes. Cell Counting Kit-8 (CCK-8; Beyotime, Jiangsu, China) assays were performed with ten replicates and Cell-LightTM EdU DNA Cell Proliferation (EdU; Ribobio, Guangzhou, China) assays were carried out in triplicate according to the manufacturer’s instructions respectively.
Mitomycin C sensitivity assay
KGN cells were seeded in 6-well plates and cultured 48 h after transfection or infection. Cells were harvested immediately or exposed to 0.1 ng/μl mitomycin C (MMC; Melonepharma) for 6 h to induce DNA damage, followed by harvested immediately or after recovery for 2 and 4 h in culture medium at 37 ℃. DNA damage marker γH2AX was detected in KGN cells with or without MMC treatment and recovery by western blot.
Clonogenic survival
HeLa cells were cultured with siRNA-Mix of PARP1/XRCC6 for 48 h. Then cells were exposed to 0.05 ng/μl mitomycin C (MMC; Melonepharma) for 6 h, followed by recovery for 2 h. The cells were harvested and counted, and reseeded into a new 6-well plate by 500 cells/well (three wells for duplicate) and incubated for 12–14 days. Colonies were stained with crystal violet (Beyotime, Jiangsu, China) and counted. Data were expressed as percent survival relative to the control: [(average treated count)/(average control count)]x100%.
Statistical analysis
All statistical analyses were performed with the use of SPSS 21.0 (IBM) and GraphPad Prism 5. Data normality were assessed by Shapiro–Wilk’s test. Continuous data in normality distribution were presented as mean ± SD and determined with the two-tailed Student’s t test; otherwise data were expressed as median (IQR) and compared by two-tailed Mann–Whitney U-test. A P value of <0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
References
Nelson, L. M. Clinical practice. Primary ovarian insufficiency. N. Engl. J. Med. 360, 606–614 (2009).
European Society for Human Reproduction and Embryology (ESHRE) Guideline Group on POI, Webber, L. et al. ESHRE guideline: management of women with premature ovarian insufficiency. Hum. Reprod. 31, 926–937 (2016).
Wu, X. et al. Impact of premature ovarian failure on mortality and morbidity among Chinese women. PLoS ONE 9, e89597 (2014).
Welt, C. K. Primary ovarian insufficiency: a more accurate term for premature ovarian failure. Clin. Endocrinol. 68, 499–509 (2008).
Knauff, E. A. et al. Anti-Mullerian hormone, inhibin B, and antral follicle count in young women with ovarian failure. J. Clin. Endocrinol. Metab. 94, 786–792 (2009).
De Vos, M., Devroey, P. & Fauser, B. C. Primary ovarian insufficiency. Lancet 376, 911–921 (2010).
Jorgez, C. J., Klysik, M., Jamin, S. P., Behringer, R. R. & Matzuk, M. M. Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol. Endocrinol. 18, 953–967 (2004).
Edson, M. A., Nagaraja, A. K. & Matzuk, M. M. The mammalian ovary from genesis to revelation. Endocr. Rev. 30, 624–712 (2009).
Matsuda, F., Inoue, N., Manabe, N. & Ohkura, S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 58, 44–50 (2012).
Dong, J. et al. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 383, 531–535 (1996).
Lechowska, A. et al. Premature ovarian failure in nobox-deficient mice is caused by defects in somatic cell invasion and germ cell cyst breakdown. J. Assist. Reprod. Genet. 28, 583–589 (2011).
Sirotkin, A. V., Ovcharenko, D., Grossmann, R., Lauková, M. & Mlyncek, M. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J. Cell Physiol. 219, 415–420 (2009).
Imbar, T. & Eisenberg, I. Regulatory role of microRNAs in ovarian function. Fertil. Steril. 101, 1524–1530 (2014).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Rah, H. et al. Association of miR-146aC>G, miR-196a2T>C, and miR-499A>G polymorphisms with risk of premature ovarian failure in Korean women. Reprod. Sci. 20, 60–68 (2013).
Pan, H. et al. The miR-449b polymorphism, rs10061133 A>G, is associated with premature ovarian insufficiency. Menopause 23, 1009–1011 (2016).
Yang, X. et al. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction 144, 235–244 (2012).
Zhang, Q. et al. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS ONE 8, e59667 (2013).
Dang, Y. et al. MicroRNA-22-3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertil. Steril. 103, 802–807.e1 (2015).
Chen, X., Xie, M., Liu, D. & Shi, K. Down regulation of microRNA-146a inhibits ovarian granulosa cell apoptosis by simultaneously targeting interleukin-1 receptor-associated kinase and tumor necrosis factor receptor-associated factor 6. Mol. Med. Rep. 12, 5155–5162 (2015).
da Rocha, S. T., Edwards, C. A., Ito, M., Ogata, T. & Ferguson-Smith, A. C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316 (2008).
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 (2010).
Gardiner, E. et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol. Psychiatry 17, 827–840 (2012).
Labialle, S. et al. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 33, 2216–2230 (2014).
Khan, S. et al. MiR-379 regulates cyclin B1 expression and is decreased in breast cancer. PLoS ONE 8, e68753 (2013).
Gururajan, M. et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transitionand bone metastasis of prostate cancer. Clin. Cancer Res. 20, 6559–6569 (2014).
Chen, J. S. et al. MicroRNA-379-5p inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in hepatocellular carcinoma. Cancer Lett. 375, 73–83 (2016).
AlAsiri, S. et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J. Clin. Invest. 125, 258–262 (2015).
Dou, X. et al. Minichromosome maintenance complex component 8 mutations cause primary ovarian insufficiency. Fertil. Steril. 106, 1485–1489.e2 (2016).
Wood-Trageser, M. A. et al. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am. J. Hum. Genet. 95, 754–762 (2014).
Desai, S. et al. MCM8 and MCM9 nucleotide variants in women with primary ovarian insufficiency. J. Clin. Endocrinol. Metab. 102, 576–582 (2017).
Qin, Y. et al. CSB-PGBD3 mutations cause premature ovarian failure. PLoS Genet. 11, e1005419 (2015).
Guo, T. et al. Mutations in MSH5 in primary ovarian insufficiency. Hum. Mol. Genet. 26, 1452–1457 (2017).
He, C. et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet. 41, 724–728 (2009).
He, C. et al. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum. Genet. 128, 515–527 (2010).
Stolk, L. et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 44, 260–268 (2012).
Day, F. R. et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat. Genet. 47, 1294–1303 (2015).
Rouleau, M., Patel, A., Hendzel, M. J., Kaufmann, S. H. & Poirier, G. G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 (2010).
Celli, G. B., Denchi, E. L. & de Lange, T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8, 885–890 (2006).
Jiang, L. et al. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J. Clin. Endocrinol. Metab. 100, E729–E738 (2015).
Gao, F. et al. Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Hum. Mol. Genet. 23, 333–341 (2014).
Titus, S. et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci. Transl. Med. 5, 172ra21 (2013).
Oktay, K., Turan, V., Titus, S., Stobezki, R. & Liu, L. BRCA mutations, DNA repair deficiency, and ovarian aging. Biol. Reprod. 93, 67 (2015).
Zhang, D. et al. Increased DNA damage and repair deficiency in granulosa cells are associated with ovarian aging in rhesus monkey. J. Assist. Reprod. Genet. 32, 1069–1078 (2015).
Bürkle, A., Beneke, S. & Muiras, M. L. Poly(ADP-ribosyl)ation and aging. Exp. Gerontol. 39, 1599–1601 (2004).
Bai, P. & Cantó, C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 16, 290–295 (2012).
Shimizu, I., Yoshida, Y., Suda, M. & Minamino, T. DNA damage response and metabolic disease. Cell Metab. 20, 967–977 (2014).
Downs, J. A. & Jackson, S. P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5, 367–378 (2004).
Ménissier de Murcia, J. et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 (2003).
Piskunova T. S. et al. Deficiency in poly (ADP-ribose) polymerase-1 (PARP-1) accelerates aging and spontaneous carcinogenesis in mice. Curr. Gerontol. Geriatr. Res. 2008, 754190 (2008).
Gu, Y. et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7, 653–665 (1997).
Li, H., Vogel, H., Holcomb, V. B., Gu, Y. & Hasty, P. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol. Cell Biol. 27, 8205–8214 (2007).
Ma, T. et al. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. J. Appl. Genet. 52, 481–486 (2011).
Tesfaye, D. et al. Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol. Reprod. Dev. 76, 665–677 (2009).
da Silveira, J. C., Veeramachaneni, D. N., Winger, Q. A., Carnevale, E. M. & Bouma, G. J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol. Reprod. 86, 71 (2012).
Jiao X. et al. Premature ovarian insufficiency: phenotypic characterization within different etiologies. J. Clin. Endocrinol. Metab. https://doi.org/10.1210/jc.2016-3960 (2017)..
Xu, X. et al. Impaired telomere length and telomerase activity in peripheral blood leukocytes and granulosa cells in patients with biochemical primary ovarian insufficiency. Hum. Reprod. 32, 201–207 (2017).
Nishi, Y. et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 142, 437–445 (2001).
Patel, A. G., Sarkaria, J. N. & Kaufmann, S. H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl Acad. Sci. USA 108, 3406–3411 (2011).
Ström, C. E., Johansson, F., Uhlén, M., Szigyarto, C. A., Erixon, K., Helleday, T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 39, 3166–3175 (2011).
Acknowledgements
The authors thank all participants. We thank Prof. Runsheng Chen and colleagues at Key Laboratory of RNA Biology of CAS, Institute of Biophysics, Chinese Academy of Sciences, China for their kind assists in microarrays data analyses. This work was supported by the National Natural Science Foundation of China (81522018, 81471509, 81701406, 81571406, and 81671413), the State Key Program of National Natural Science Foundation of China (81430029), National Research and Development Plan (2016YFC1000604), National Key Research & Developmental Program of China (2017YFC1001100).
Author information
Authors and Affiliations
Contributions
Y.Q. and Z.-J.C. designed the research; Y.D., X.W., X.Z., S.Z., and J.M. performed the research; Y.D. and Y.H. analyzed the data; and Y.D., X.W., Y.Q., and Z.-J.C. wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by E. Candi
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dang, Y., Wang, X., Hao, Y. et al. MicroRNA-379-5p is associated with biochemical premature ovarian insufficiency through PARP1 and XRCC6. Cell Death Dis 9, 106 (2018). https://doi.org/10.1038/s41419-017-0163-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-017-0163-8
This article is cited by
-
Comprehensive analysis of lncRNA-miRNA-mRNA ceRNA network and key genes in granulosa cells of patients with biochemical primary ovarian insufficiency
Journal of Assisted Reproduction and Genetics (2024)
-
Granulosa Cells-Related MicroRNAs in Ovarian Diseases: Mechanism, Facts and Perspectives
Reproductive Sciences (2024)
-
X-ray cross-complementing family: the bridge linking DNA damage repair and cancer
Journal of Translational Medicine (2023)
-
Identification of transcriptome characteristics of granulosa cells and the possible role of UBE2C in the pathogenesis of premature ovarian insufficiency
Journal of Ovarian Research (2023)
-
MicroRNA-338-3p helps regulate ovarian function by affecting granulosa cell function and early follicular development
Journal of Ovarian Research (2023)