Abstract
Neonatal mammalian heart maintains a transient regeneration capacity after birth, whereas this regeneration ability gradually loses in the postnatal heart. Thus, the reactivation of cardiomyocyte proliferation is emerging as a key strategy for inducing heart regeneration in adults. We have reported that a highly conserved long noncoding RNA (lncRNA) LncDACH1 was overexpressed in the failing hearts. Here, we found that LncDACH1 was gradually upregulated in the postnatal hearts. Cardiac-specific overexpression of LncDACH1 (TG) in mice suppressed neonatal heart regeneration and worsened cardiac function after apical resection. Conversely, in vivo cardiac conditional knockout of LncDACH1 (CKO) and adenovirus-mediated silencing of endogenous LncDACH1 reactivated cardiomyocyte-proliferative potential and promoted heart regeneration after myocardial infarction (MI) in juvenile and adult mice. Mechanistically, LncDACH1 was found to directly bind to protein phosphatase 1 catalytic subunit alpha (PP1A), and in turn, limit its dephosphorylation activity. Consistently, PP1A siRNA or pharmacological blockers of PP1A abrogated cardiomyocyte mitosis induced by LncDACH1 silencing. Furthermore, LncDACH1 enhanced yes-associated protein 1 (YAP1) phosphorylation and reduced its nuclear translocation by binding PP1A. Verteporfin, a YAP1 inhibitor decreased LncDACH1 silencing-induced cardiomyocyte proliferation. In addition, targeting a conserved fragment of LncDACH1 caused cell cycle re-entry of human iPSC-derived cardiomyocytes. Collectively, LncDACH1 governs heart regeneration in postnatal and ischemic hearts via regulating PP1A/YAP1 signal, which confers a novel therapeutic strategy for ischemic heart diseases.
Similar content being viewed by others
Introduction
Myocardial infarction (MI) is one of the leading causes of morbidity and mortality in patients suffering from cardiovascular diseases worldwide. Due to the massive loss of cardiomyocytes, the infarct heart undergoes post-infarct remodeling and degenerates eventually to heart failure [1]. One major strategy to overcome long-term health threatening of cardiac infarction is to remedy the infarcted heart by either reducing the loss of cardiomyocytes or increasing the number of cardiomyocytes exogenously or endogenously. Recently, studies reveal that the neonatal cardiomyocytes, and to some extent, adult cardiomyocytes, can obtain proliferative and regenerative properties under certain circumstances, highlighting the potential of repairing infarct heart by enhancing cardiac regeneration endogenously [2,3,4].
Great efforts have been made to identify novel molecules and molecular mechanisms that hold the therapeutic potential for cardiac infarction-related cell loss [5,6,7,8,9,10,11]. Transcriptional factors, such as Meis1, Pitx2, Tbx20, and Gata4, have been shown to play a pivotal role in regulating the proliferative capacity of mammalian cardiomyocytes [2, 4, 5]. The Hippo/YAP1 signaling pathway is known to regulate division of cardiomyocytes, and the regeneration of damaged myocardium through multiple transcriptional mechanisms [6]. Moreover, environmental hypoxia or reduced oxygen metabolism in the adult mammalian heart can induce cell cycle re-entry of cardiomyocytes, which contributes to the improvement of cardiac functions following MI [12, 13]. Despite the growing attention paid to cardiomyocyte proliferation in cardiac repair, the precise mechanisms that govern cardiomyocyte-regenerative capacity are not thoroughly understood and remain a central subject in the field of cardiac biology research.
Noncoding RNAs (ncRNAs) are RNAs that do not encode protein, while they regulate gene expression with multiple mechanisms, and exert a wide range of biological functions such as apoptosis, proliferation, and differentiation [14,15,16]. Emerging studies have demonstrated that both small ncRNAs (miRNAs) and long ncRNAs (lncRNAs) play vital roles in tissue development, cancer, neurodegenerative diseases, and heart disorders [10, 15,16,17]. LncRNAs have recently been recognized as critical regulators of development, physiological function, and pathological processes of the heart [18]. For example, Braveheart plays an important role in cardiomyocyte differentiation of pluripotent stem cells [18]. CARMEN, a human super enhancer-associated lncRNA, acts as a crucial regulator of cardiac specification and differentiation of cardiac precursor cells [19]. The expression level of CARMEN is significantly upregulated in hypertensive patients, and in the mouse with cardiac pathological remodeling [20]. The lateral mesoderm-specific lncRNA Fendrr regulates heart and body wall development by targeting several transcription factors such as Gata6 and Nkx2–5 in the mouse [21]. The lncRNA maternally expressed gene 3 (Meg3) was also found to take part in the development of cardiac fibrosis and diastolic dysfunction after chronic pressure overload [22]. Wang et al. demonstrated that a heart-enriched lncRNA, named cardiac-hypertrophy-associated epigenetic regulator (Chaer), is necessary for stress-induced cardiac hypertrophy as an epigenetic checkpoint [23]. Recently, we reported that LncDACH1 upregulation impaired cardiac function by promoting ubiquitination-related degradation of SERCA2a [24]. Nevertheless, the current knowledge about the regulation of lncRNAs on cardiac disorders is still rather limited, especially the potential roles of lncRNAs in regulating cardiomyocyte proliferation and cardiac repair.
In this study, we observed that the highly conserved lncRNA LncDACH1 was gradually upregulated after birth, and contributed to the loss of cardiomyocyte proliferation and regenerative capacity. We further identified that LncDACH1 acts as a protein phosphatase 1 catalytic subunit alpha (PP1A) inhibitor to regulate the Hippo/YAP1 pathway, and thus controls the mitosis of cardiomyocytes and myocardial regeneration. Our observations raise the possibility that targeting LncDACH1 reactivates cardiomyocyte proliferation and promotes heart regeneration after injury in mammals.
Materials and methods
Ethical approval
Our animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Harbin Medical University. The procedures conform to the Guide for the Care and Use of Laboratory Animals, published by the U.S. National Institutes of Health. C57BL/6 mice were purchased from the Experimental Animal Center of the Affiliated Second Hospital of Harbin Medical University (Harbin, China). Food and water were freely available throughout the experiments.
Human plasma was obtained from people with or without MI. After a normal medical examination, the leftover blood samples were used to detect the expression of LncDACH1. The study protocol was approved by the Ethics Committee of Harbin Medical University (HYDEY2018-184), and written informed consent was obtained from all participants. All procedures were carried out in accordance with the Declaration of Helsinki (2013).
Generation of Myh6-LncDACH1 transgenic mice
Myh6-LncDACH1 transgenic mice on a C57BL/6 background were purchased from Beijing Biocytogen Co. Ltd. (Beijing, China). Briefly, the sequence of LncDACH1 was cloned into the murine Myh6 promoter expression vector, and the obtained DNA fragment containing LncDACH1 driven by Myh6 promoter was microinjected into zygotes. The zygotes were then implanted as a group of 10–15 bilaterally into the oviduct of pseudopregnant mice. Animal genotype was verified by PCR on tail genomic DNA. The primer pairs used are forward primer TACCAAACCAATTGAATTGCGCTGATGCC-3 and reverse primer CCCCATCCCTGCAGGCATTC. To maintain an isogenic strain, mice were propagated as heterozygotes by breeding with WT C57BL/6 mice. LncDACH1 overexpression in the heart was verified by qRT-PCR.
Generation of cardiac-specific LncDACH1 conditional knockout (CKO) mice
The αMHC-MerCreMer mice (stock number 005657) were obtained from the Jackson laboratory. LncDACH1fl/fl mice were generated by Beijing Biocytogen Co. Ltd. (Beijing, China). The mice were maintained on a C57BL/6 J genetic background. To generate mice with cardiomyocyte-specific LncDACH1 gene knockout, LncDACH1fl/fl mice were crossed with Cre transgenic mice. The adult Cre + LncDACH1fl/fl (CKO) mice were injected with tamoxifen (Sigma) at 80 mg/kg body weight intraperitoneally once a day for five consecutive days. The control group of the conditional knockout mice was Cre-LncDACH1fl/fl (Control) mice, which lack the Cre transgene, and also received tamoxifen injections. The genotype of Cre and LncDACH1fl/fl mice was verified by PCR on tail genomic DNA. The primer pairs of LncDACH1fl/fl used in this study are LncDACH1 loxP–F: CCAGAAGCACCCAGGACATTGTTGT, LncDACH1 loxP–R: ACATCACAGAGCCACTGTAAGGAGTT. The primer pairs of Cre are wt-R (28887): CCAACTCTTGTGAGAGGAGCA, mutant-R (28888): CCAGCATTGTGAGAACAAGG, and common-F (28886): TCTATTGCACACAGCAATCCA. The WT and floxed PCR products correspond to 306 and 394 bp, respectively. The expression loss of LncDACH1 in the heart induced by tamoxifen was verified by qRT-PCR.
Construction of adenovirus-carrying LncDACH1
Adenovirus vectors carrying LncDACH1 (Adv-LncDACH1), or a short RNA fragment for silencing LncDACH1 (Adv-shLncDACH1) and a CAG promoter, were constructed by Genechem Co. Ltd. (Shanghai, China). Briefly, the adenoviral shuttle vector carrying the gene of interest was co-transfected into HEK293 cells, with an auxiliary packaging plasmid carrying the majority of the genome of the adenovirus. The Cre/loxP recombinase system recombined the recombinant adenovirus. Then the test genes were integrated into the E1- and E3-deleted adenoviral genome. Adenovirus was then purified by using the Adeno-X™ Virus Purification Kit (BD Biosciences, Clontech).
Apical resection
Apical resection was performed on neonatal mice at postnatal day 1 (P1). Littermate mice were randomly selected from the cage, and assigned to different control and experimental subgroups. Randomization was done by selecting animals of similar age and weight. Neonates were anesthetized by cooling on an ice bed for 4 min. Lateral thoracotomy at the fourth intercostal space was performed by blunt dissection of the intercostal muscles following skin incision. Iridectomy scissors were used to resect the apex of P1 hearts, until the left ventricular chamber was exposed. Following apical resection, adenoviral LncDACH1 (2 × 109 PFU/mL) or its relevant control were injected into the myocardium. Then their thoracic wall incisions were sutured with 7-0 nonabsorbable silk suture, and the skin wounds were closed by using skin adhesive. Sham-operated mice underwent the same procedure without apical resection. The neonates were then placed under a heat lamp, and warmed for several minutes until recovery. Hearts were collected 14 or 21 days after resection.
Myocardial infarction (MI)
MI was established in juvenile (P7) or male adult-stage (6–8 weeks old) C57BL/6 mice. Juvenile mice were anaesthetized by cooling on an ice bed for 5 min. The adult mice received tracheal intubation and were ventilated with 3% isoflurane for induction and 2% isoflurane for maintenance of anesthesia. Then, the left anterior descending coronary artery was ligated with 7-0 prolene suture, and 2 × 109 adenoviral LncDACH1 shRNA or NC was injected into the myocardium following MI. After completion of the surgery, the chest was closed. Mice were then warmed for several minutes until recovery. Hearts were collected 14 days after the operation for further analyses.
Echocardiography
Cardiac function was evaluated by two-dimensional echocardiography in conscious mice. Echocardiography was performed by Vevo 1100 VisualSonics devices (VisualSonics, Toronto, ON, Canada). Fractional shortening (FS) and ejection fraction (EF) were measured by using M-mode images taken from the parasternal short-axis view. Echocardiography data were analyzed by investigators blinded to treatment and genotype. The average of at least three measurements was used for each single data point.
Wheat germ agglutinin (WGA)
Hearts were harvested and snap-frozen in liquid nitrogen with optimal cutting temperature (OCT) compound. Hearts were sliced into 6-μm sections. Then sections were fixed in acetone followed by washing with PBS, blocked with goat serum, and then incubated with wheat germ agglutinin (WGA) conjugated with Alexa 488 (#W11261; Thermo Fisher, Carlsbad, CA, USA). Cell size was measured by using Image J software. The observers and statisticians were blinded from group identities.
Histological analysis
After surgery and measurements, hearts were collected and fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 6-µm intervals. H&E staining and Masson’s trichrome staining were performed according to standard procedures. The observers and statisticians were blinded to the group identities.
Immunohistochemistry
Hearts were harvested and snap-frozen in liquid nitrogen with OCT compound, and cut into 6-μm sections. The sections were fixed in acetone followed by washing with PBS, blocked with goat serum, and then incubated with primary antibodies phospho-Histone H3 (1:500; #06-570, Millipore, Billerica, MA, USA), Aurora B (1:500; ab2254, Abcam, Cambridge, MA, UK), and α-actinin (1:400; ab9465, Abcam, Cambridge, MA, UK) overnight. The preparations were subsequently incubated with secondary antibodies goat anti-mouse IgG (H&L) (Alexa Fluor 488; 1:400; ab150113, Abcam) or goat anti-rabbit IgG H&L (Alexa Fluor 594; 1:400; ab150080, Abcam). DAPI staining was performed to visualize nuclei. Images were acquired by using a confocal laser scanning microscope (BX53; Olympus, Japan). The observers and statisticians were unaware of the sample group allocation during the experiment and when assessing its outcome.
Culture of human-induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM)
Undifferentiated hiPS cells (AC-iPSC) were maintained on Matrigel-coated plates in an E8 medium (CA1001500, CELLAPY), which was changed every day. When confluence reached 60–70%, hiPSCs were washed with PBS once and then incubated with 0.5 mM EDTA for 1 min at 37 °C. The cells were washed gently four times with E8 medium, and replanted in Matrigel-coated plates to 1:10 dilutions. For monolayer cardiomyocyte differentiation, 70–80% confluent hiPSCs were cultured with differentiation basal medium comprising RPMI1640 medium (C11875500BT, Thermo Fisher Scientific) and B27 minus insulin (A1895601, Thermo Fisher Scientific). The hiPSCs were incubated in differentiation basal medium added with CHIR-99021 (HY-10182, MCE) for 1 day and Wnt-C59 (S7037, Selleck Chemicals) for 2 days. Finally, the medium was aspirated, and the cells were subsequently fed every 1–2 days in RPMI1640 basal medium containing B27 (17504044, Thermo Fisher Scientific). Beating cells were observed at day 8–9 after differentiation.
Isolation of cardiomyocytes from neonatal mice
Cardiomyocytes were isolated from neonatal mice (postnatal day 1–2). Briefly, hearts were harvested from neonatal mice and rinsed using PBS. Then, hearts were cut into pieces and digested using trypsin (Bl121001; Delta). The supernatant was collected and centrifuged to separate the cells. The cells were suspended in DMEM with 4.5 g/L glucose (Life Technologies), supplemented with 10% FBS, 100 U/ml of penicillin, and 100 mg/ml of streptomycin (Sigma), and then seeded onto 100-mm plastic dishes in 5% CO2 and humidified atmosphere at 37 °C for 1.5 h. Then, the cells suspended in DMEM were seeded onto 0.1% gelatin-coated glass-bottom cell culture dishes or six-well plates with a suitable density in 5% CO2 and humidified atmosphere at 37 °C for 48 h.
Construction of LncDACH1 plasmid and truncated LncDACH1 fragment plasmid
LncDACH1 cDNA and other fragments of cDNA were inserted into the pCDNA3.1, respectively. The sequence of LncDACH1 was divided into five fragments. First, we cut off the first 417 sequences of the entire sequences and constructed LncDACH1-b (418–2085), and then another 417 sequences were cut off to generate LncDACH1-c (835–2085). Likely, other truncated LncDACH1 fragments LncDACH1-d (1251–2085), LncDACH1-e (835–1668), LncDACH1-f (835–1251), and LncDACH1 (835–1251) mut were constructed.
Transfection
The isolated cells were cultured in six-well plates or glass-bottom cell culture dishes (801001, NEST Biotechnology) at a 70–80% density per well. After 48 h, cells were transfected with LncDACH1 plasmid (Genechem, Shanghai, China) or LncDACH1 siRNA (GenePharma, Shanghai, China) using Lipofectamine 3000 or Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Cardiomyocytes following transfection were cultured in a humidified atmosphere at 37 °C in 5% CO2 for 48 h. Then cells were collected for further analysis. The expression of LncDACH1 in cardiomyocytes after transfection was detected by using qRT-PCR. The target sequences of LncDACH1 siRNA are sense 5′-CACCCUAGUUUCUUUCAAUTT-3′ and antisense 5′-AUUGAAAGAAACUAGGGUGTT-3′.
Pharmacological reagent treatment
Pharmacological inhibitors of protein phosphatase 1, calyculin A (Hy-18983), were purchased from MedChem Express, and okadaic acid (07760) from Sigma. The YAP1 inhibitor, verteporfin (HY-B0146) was obtained from MedChem Express. Cardiomyocytes were incubated with 25 nM calyculin A for 30 min, 1 μM okadaic acid for 3 h, and 2 μM verteporfin for 24 h following transfection with LncDACH1 siRNA for 48 h.
EdU incorporation assay
After transfection with the appropriate reagents for 48 h, and incubation with EdU for 12 h, cardiomyocytes were fixed and labeled by using In Cell-Light EdU Apollo 567 in Vitro Kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions.
Immunocytochemistry
After transfection, cells were fixed with 4% paraformaldehyde (P1110; Solarbio) for 15 min, and permeabilized with 0.4% Triton X-100 (T8200; Solarbio) in PBS for 60 min, followed by 60 min of blocking in goat serum (AR0009; Boster). Cells were then stained with primary antibodies against phospho-Histone H3 (1:500; #06-570, Millipore, Billerica, MA, USA), Aurora B (1:500; ab2254, Abcam), α-actinin (1:400; ab9465, Abcam, Cambridge, MA, UK), or YAP1 (1:200; 13584-1-AP, Proteintech) at 4 °C overnight, followed by incubation with a secondary antibody conjugated to Alexa Fluor-488 or Alexa Fluor-594 for 1 h at room temperature. Cells were counterstained with DAPI (C0065, Solarbio) to label nuclei. The EdU+, pH3+ and Aurora B+ cardiomyocytes were then evaluated by a confocal laser scanning microscope (FV10i; Olympus, Japan).
Quantitative real-time PCR (qRT-PCR)
For transfection efficiency analysis, RNAs were extracted after varying treatments by using Trizol reagent (Invitrogen, Carlsbad, CA), and reverse transcribed into cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol using a random primer. Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on ABI 7500 fast Real-Time PCR system (Applied Biosystems, USA). GAPDH or 18s were used as internal controls. Expression levels of LncDACH1 mRNA were calculated by the relative quantitative 2−▵▵CT method. The specific sequences of primer pairs for LncDACH1 used are Mus LncDACH1 Forward: 5′-AAGATAGGATGTTGGGGCAG-3′, Mus LncDACH1 Reverse: 5′-ACCATAGCACAAACACTTCC-3′, Hsa LncDACH1 Forward: 5′-TTGCAACAGCCAAGTTCATGC-3′, Hsa LncDACH1 Reverse: 5′-CTGTCCTTGATCAGCTTCCTCC-3′, and Mus DACH1 Forward: 5′-AACCGCAAGAGACAGCATCG-3′, Mus DACH1 Reverse: 5′-GGACAGGCCATCAGGAAACAG-3′.
Western blot
The protein samples (80 μg) were loaded onto SDS-PAGE gels for protein electrophoresis, and then transferred onto nitrocellulose membranes by an electroblot apparatus. Antibodies against PP1A (ab52619; Abcam), Tropomodulin (NBP2-00955; Novusbio), DACH1 (OM109425; Omnimabs) and CTNND1 (HPA015955; Sigma) were used as primary antibodies. Mouse or rabbit IgG antibodies coupled to horseradish peroxidase were used as secondary antibodies. GAPDH (AC033; ABclonal) and beta-actin (AC026; ABclonal) were used as loading control. Quantification of Western blot was performed by Odyssey (LI-COR Biosciences, Lincoln, NE, USA).
Fluorescent In Situ Hybridization (FISH)
RNA-FISH assay was performed with a Fluorescent In Situ Hybridization (FISH) Kit (RiboBio, Guangzhou, China). Briefly, cardiomyocytes were fixed in 4% formaldehyde at room temperature for 10 min and permeabilized with 0.5% Triton-100 in PBS at 4 °C for 30 min after washed with PBS solution. Then, cells were prehybridized at 37 °C for 30 min, followed by hybridization using lncRNA probe in hybridization solution at 37 °C overnight. After hybridization, cells were washed with prewarmed wash buffer and PBS solution for six times. Finally, cells were counterstained with DAPI, and visualized using a confocal laser scanning microscope (FV10i; Olympus, Japan).
RNA binding protein immunoprecipitation assay (RIP)
Cardiomyocytes were scraped off the dish using a cell lifter after washing with ice-cold PBS, and centrifuged at 1500 rpm/min for 5 min. The cell pellet was collected and resuspended with lysis buffer (150 mmol/L KCl, 25 mmol/L Tris-Cl, pH 7.4, 5 mmol/L EDTA, 0.5% NP-40, 0.5 mmol/L DTT, 100 U/mL RNase inhibitor (Sigma), and 1× protease inhibitor cocktail) followed by incubation on ice for 5 min. After centrifugation at 14,000 rpm/min for 5 min, lysate was incubated with protein A/G magnetic beads that have been pre-hybridized with PP1A antibody at 4 °C on a rotating wheel for 3 h. The beads were collected and purified using protease K, and then the RNAs in the supernatant were isolated using TRIZOL reagent. PCR was used to detect the enriched RNAs. Normal IgG was used as negative control.
RNA–protein pull down and mass spectrometry assay
LncDACH1 plasmid DNA was linearized and then transcribed into RNAs using MEGAscript® Kit (Ambion). The RNAs were labeled with biotin by Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo). Briefly, the labeled RNAs were incubated with streptavidin magnetic beads on a shaker at room temperature for 30 min; then the bead-bound RNAs were collected and incubated with RNA–protein-binding reaction mixture (10 μL of 10× protein–RNA-binding buffer, 30 μL of 50% glycerol, and 150 μg of lysate, and add nuclease-free water to 100 μL). The eluted RNA binding protein was then analyzed by mass spectrometry assay (Aksomic) and Western blot. The antisense of LncDACH1 was used as negative control (NC).
PP1A activity detection assay
PP1A activity was detected using PP1A activity detection assay (HL50313.1; Haling, Shanghai, China) according to the manufacturer’s instructions. The PP1A activity was measured at 660 nm, and calculated using the formula PP1A activity = {[C(sample) − blank)]*10* sample dilution multiple}/reaction time/protein concentration.
Antibodies and reagents
All antibodies were purchased commercially: anti-phospho-Histone H3 (1:500; #06-570, Millipore, Billerica, MA, USA), anti-Aurora B (1:500; ab2254, Abcam), anti-α-actinin (1:400; ab9465, Abcam, Cambridge, MA, UK), anti-YAP1 (1:200; 13584-1-AP, Proteintech), anti-PP1A (ab52619, Abcam), anti-Tropomodulin (NBP2-00955, Novusbio), anti-CTNND1 (HPA015955, Sigma), and anti-GAPDH (AC033, ABclonal). Secondary antibodies for immunofluorescence staining were goat anti-mouse IgG (H&L) (Alexa Fluor 488; 1:400; ab150113, Abcam), goat anti-rabbit IgG H&L (Alexa Fluor 594; 1:400; ab150080, Abcam). Wheat germ agglutinin (WGA) conjugated with Alexa 488 (#W11261; Thermo Fisher, Carlsbad, CA, USA). Reagents used in this study: B27 minus insulin (A1895601, Thermo Fisher Scientific), CHIR-99021 (HY-10182, MCE), and Wnt-C59 (S7037, Selleck Chemicals). Calyculin A (Hy-18983) was purchased from MedChem Express, and okadaic acid (07760) from Sigma. Verteporfin (HY-B0146) was obtained from MedChem Express.
Statistics
Sample sizes, as indicated in figure legends, were selected based on effect size and availability as per usual standard. Data are expressed as mean ± SEM. Data distribution was evaluated by the Kolmogorov–Smirnov test. An F-test was used to evaluate homogeneity of variance. Normally distributed data with only one variable were analyzed by parametric analysis: an unpaired (two-tailed) Student’s t test (with Welch correction when variance was unequal) for two groups and one-way analysis of variance (ANOVA) with post hoc Dunnett’s/Newman–Keuls test for more than two groups. Non-normally distributed data with only one variable were analyzed by nonparametric analysis: Mann–Whitney U test (two-tailed) for two groups and one-way analysis of variance (ANOVA) with a Kruskal–Wallis test for more than two groups. Data with more than one variable were evaluated by two-way ANOVA, with Newman–Keuls’s Multiple Comparison tests. Data were analyzed using Graphpad Prism software (version 5.0, GraphPad Software Inc., San Diego, California, USA). P < 0.05 was considered statistically significant.
Results
LncDACH1 overexpression renders loss of regenerative capacity in neonatal hearts
LncRNAs have emerged as new and critical regulators of a variety of heart diseases. Lately, we discovered a highly conserved lncRNA with 2085-nt in length was associated with the development of heart failure [24]. It is located in the first intron of DACH1 gene in mouse (named as LncDACH1) (Supplementary Fig. 1). Interestingly, we found that LncDACH1 was markedly upregulated during postnatal heart development (Supplementary Fig. 2), which is coincident with the reported loss of myocardial regenerative capacity. Meanwhile, the expression level of LncDACH1 in the heart tissue and plasma of MI mice was higher than that of sham mice (Supplementary Fig. 3). The translational analysis with RegRNA 2.0 showed that LncDACH1 possesses no major open-reading frames (ORFs) with translational propensity (Supplementary Fig. 4a). The FISH assay demonstrated that LncDACH1 was mainly distributed in the cytoplasm of cardiomyocytes (Supplementary Fig. 4b).
We then explored the potential role of LncDACH1 in cardiomyocyte proliferation and heart regeneration by using both gain- and loss-of-function approaches. First, we generated cardiac-specific LncDACH1 transgenic (TG) mice and evaluated its regenerative capacity on neonatal hearts. In 1-day old newborn mice, the heart to body weight ratio was of no significant difference between WT and TG mice (Fig. 1a, Supplementary Fig. 5a), while the number of mitosis marker phospho-histone H3 (pH3) and cytokinesis marker Aurora B positive cells was significantly reduced in TG mice as compared with WT mice (Fig. 1b). Besides, the surface size of cardiomyocytes was increased in the heart of TG mice compared with WT mice (Fig. 1c), indicating that LncDACH1 overexpression inhibits cardiomyocyte proliferation and mitosis in mice. We further explored the regenerative capacity of neonatal hearts in response to apex resection. In sham groups, like 1-day old TG mice, 14-day old TG mice also demonstrated reduced cardiac mitosis and increased cell size relative to WT controls (Fig. 2a). In apex resection groups, the number of pH3 positive and Aurora B positive cells was decreased, and cell size was increased in LncDACH1 TG mice compared with WT controls (Fig. 2a). Consistently, the cardiac function of LncDACH1 TG mice of both the sham and resection groups was markedly impaired relative to WT controls (Fig. 2b, c). In addition, intramyocardial injection of adenovirus vector carrying LncDACH1 gene (Adv-LncDACH1) was also used to achieve LncDACH1 overexpression in vivo (Supplementary Fig. 5b). Consistent with LncDACH1 transgenic overexpression mice, adenovirus-mediated LncDACH1 overexpression at P1 of neonatal heart significantly inhibited the mitosis and cytokinesis of cardiac myocytes, increased cell size and impaired cardiac function in both sham and apex resection hearts (Fig. 3a–c). In addition, the percentage of cardiomyocytes isolated from neonatal mice with EdU, pH3 and Aurora B positive staining was significantly decreased 48 h after LncDACH1 overexpression, and the number of binucleated cardiomyocytes was also markedly increased (Supplementary Figs. 5c and 6). These results indicate that LncDACH1 overexpression inhibits cardiomyocyte proliferation and suppresses the capacity of cardiac regeneration.
a Representative images of the whole heart and H&E-stained cardiac sections of 1-day-old mice showing no change in heart/body weight between TG and WT mice. WT, n = 7; TG, n = 9. Two-tailed Student’s t test. Scale bars = 500 μm. b Heart sections from 1-day- old mice. Mitosis was visualized with an antibody specific for phospho-histone H3 (pH3); cytokinesis was visualized with an antibody specific for Aurora B. Scale bars = 20 μm. n = 5/group. Two-tailed Student’s t test with or without Welch’s correction. c Cell cross-sectional area was determined by wheat germ agglutinin (WGA). Scale bars = 20 μm. n = 5. Two-tailed Student’s t test with Welch’s correction. Data are shown as mean ± SEM, *P < 0.05, ***P < 0.001.
a Apex resection (AR) was performed on 1-day-old WT and TG mice, hearts were harvested and stained with pH3 for mitosis (n = 5, 5, 4, 4), Aurora B for cytokinesis (n = 3, 3, 3, 5), WGA for cell cross-sectional area (n = 4, 3, 3, 3), α-actinin, and DAPI 14 days post AR. Scale bars = 20 μm. One-way ANOVA followed by Newman–Keuls’s Multiple Comparison tests. b Gross morphology and H&E staining for WT and TG mice 21 days after AR. Scale bars = 500 μm. c Serial echocardiographic measurements of ejection fraction (EF) and fraction shortening (FS) (n = 4, 4, 4, 6). One-way ANOVA with Newman–Keuls’s Multiple Comparison tests. Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001.
a Representative heart section staining with pH3 for mitosis (n = 3–5), Aurora B for cytokinesis (n = 3–5), and WGA for cell cross-sectional area (n = 3/group). Neonatal mice at postnatal day 1 (P1) was subjected to apical resection surgery and injected with Adv-LncDACH1 or Adv-Control. Scale bars = 20 μm. b Gross morphology and H&E staining for the hearts with AR and Adv-LncDACH1 injection for 21 days. Scale bars = 500 μm. c Representative echocardiography, ejection fraction (EF) and fraction shortening (FS). n = 4 for Adv-Control group, n = 3 for Adv-LncDACH1 group, n = 4 for AR + Adv-Control group, n = 6 for AR + Adv-LncDACH1 group. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA followed by Newman–Keuls’s post hoc test for Multiple Comparison tests.
Cardiac conditional knockout of LncDACH1 enhances the proliferative potential of cardiomyocytes after ischemic injury
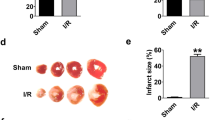
After obtaining the evidence for the negative role of LncDACH1 in cardiac regeneration with gain-of-function results as described above, we went on to verify the data with loss-of-function studies to further validate the regulation of LncDACH1 on cardiac regeneration. To this end, we generated a line of cardiac-specific LncDACH1 conditional knockout (CKO) mice and explored the reparative potential of LncDACH1 silencing after cardiac injury induced by coronary artery ligation. The cardiac conditional knockout of LncDACH1 in mice was confirmed by qRT-PCR (Supplementary Fig. 7a). Cardiac conditional knockout of LncDACH1 did not change basal cardiac function. However, it improved the cardiac function of MI mice (Fig. 4a). The infarct size of the left ventricle was smaller in LncDACH1 CKO mice than in controls (Fig. 4b). Meanwhile, a higher number of pH3 and Aurora B-positive cells were observed in LncDACH1 CKO mice than in control counterparts under both sham and MI conditions (Fig. 4c). Consistently, the surface size of ventricular myocytes was smaller in CKO mice than in controls (Fig. 4d). These data suggest that loss of LncDACH1 enhances cardiomyocyte proliferation and thus promotes cardiac regeneration after injury in mice.
a Representative echocardiography, EF, and FS for adult CKO mice. Adult mice were injected intraperitoneally once a day for five consecutive days with tamoxifen, and MI was established 7 days after tamoxifen injection. Hearts were collected 14 days after operation. n = 7 for Control group, n = 7 for LncDACH1 CKO group, n = 5 for Control + MI group, n = 4 for LncDACH1 CKO + MI group. One-way ANOVA followed by Newman–Keuls’s Multiple Comparison tests. b Representative Masson trichrome staining of heart sections from adult MI mice. Quantification of infarct scar size 14 days post MI. Scale bars = 1 mm. n = 5/group. Two-tailed Student’s t test. c Quantification of the percentage of pH3-positive cells for mitosis (n = 3–5) and Aurora B for cytokinesis (n = 4–5) in adult CKO hearts. Scale bars = 20 μm. d Cardiomyocytes cross-sectional area measured by WGA staining. Scale bars = 20 μm. n = 4/group, one-way ANOVA followed by Newman–Keuls’s Multiple Comparison tests. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001.
LncDACH1 deficiency reactivates cardiomyocyte proliferation and improves cardiac function in MI
To further verify the effects of LncDACH1 knockdown on cardiac regeneration, adenovirus carrying a LncDACH1 shRNA (Adv-shLncDACH1) was injected into the myocardium of 7-day-old mice, and cardiac reparative potential was examined 2 weeks after coronary artery ligation. As illustrated in Fig. 5a, knockdown of LncDACH1 by Adv-shLncDACH1 increased the number of pH3- and aurora B-positive cells relative to the negative control adenovirus construct. Meanwhile, Adv-shLncDACH1 treatment significantly decreased the cell size of cardiomyocytes, minimized infarct area and improved cardiac function in infarct hearts (Fig. 5a, b). These results imply that silencing of LncDACH1 enhances the proliferation of cardiomyocytes and promotes cardiac regeneration in juvenile hearts after injury. Consistently, in vitro, knockdown of LncDACH1 with its siRNA (Supplementary Fig. 7b) increased the number of pH3- and aurora B-positive cells, and reduced the number of binucleated cells in cultured mouse cardiac myocytes (Fig. 5c).
a Hearts from animals were immunostained by pH3 for mitosis (n = 5), Aurora B for cytokinesis (n = 4), and WGA for cell cross-sectional area (n = 4). Scale bars = 20 μm. b The fibrosis area was determined by Masson trichrome staining (n = 3–6) and cardiac function was determined by echocardiography (n = 5). Scale bars = 1 mm. c The proliferation ability of cardiomyocytes was shown by EdU and pH3 staining after inhibition of LncDACH1. Scale bars = 50 μm. Data are shown as mean ± SEM, n = 5 per group. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test with or without Welch’s correction.
We then explored whether the proregenerative effects of LncDACH1 knockdown are also present in the infarct hearts of adult mice. Eight-week-old mice were subjected to coronary ligation and then injected with Adv-shLncDACH1, followed by evaluation of cardiac reparative response two weeks after the operation. As shown in Supplementary Fig. 7c, the expression of LncDACH1 in the heart was successfully silenced by Adv-shLncDACH1. Knockdown of LncDACH1 increased the regenerative potential of hearts after infarction in adult mice, as indicated by increased pH3 positive and Aurora B positive cells and decreased cell size (Fig. 6a). Masson staining and echocardiographic measurements showed that the Adv-shLncDACH1 administration also reduced the infarct size of adult mice and improved cardiac function (Fig. 6b, c). These data suggest that knockdown of LncDACH1 also promotes cardiac regeneration and reactivates cardiomyocyte proliferation.
a The level of pH3 for mitosis (n = 5), Aurora B for cytokinesis (n = 4), and cross-sectional area (n = 5) in adult mice hearts 14 days post MI were determined by immunofluorescence. Scale bars = 20 μm. b Representative histological sections at day 14 after MI of adult mice through multiple levels of the heart (apex toward base) with Masson’s trichrome staining. The scar tissue is indicated in blue and the healthy myocardium in red. Bar graph shows the quantification of scar size by histology (n = 3–4). Scale bars = 1 mm. c Cardiac function by echocardiography (n = 4). Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, two-tailed Student’s t test.
LncDACH1 interacts with the PP1A
It is well documented that the function and mechanism of lncRNAs are closely correlated with their host genes. However, our data showed that LncDACH1 does not affect the expression of its host gene DACH1 (Supplementary Fig. 8), indicating LncDACH1 functions independent of DACH1. To get further insight into the molecular mechanism underlying the effects of LncDACH1 on cardiac regeneration, we performed an RNA pull-down experiment followed by protein mass spectrometry to screen for the proteins that interact with LncDACH1. As shown in Supplementary Table 1, more than thirty proteins were found to be associated with LncDACH1, among which Tropomodulin 1 (Tmod1), CTNND1 and PP1A demonstrated the higher affinities to LncDACH1 (Supplementary Table 1). However, the results from RNA pull-down and immunoblotting experiments showed that PP1A was successfully precipitated by LncDACH1 instead of Tmod1 and CTNND1 compared with the antisense (Fig. 7a). RNA immunoprecipitation study showed that PP1A precipitated more LncDACH1 than IgG control (Fig. 7b). Then, we detected whether the binding of LncDACH1 and PP1A affects the activity of PP1A. The data showed that overexpression of LncDACH1 decreased the activity of PP1A, while silencing of LncDACH1 increased its activity (Fig. 7c). These results suggest that LncDACH1 directly binds to PP1A and limits its activity in cardiomyocytes.
a PP1A was pulled down by LncDACH1. b RNA immunoprecipitation (RIP) analysis using the PP1A antibody followed by qRT-PCR for LncDACH1. Data were normalized to the corresponding normal IgG RIP groups. n = 4 per group, two-tailed Student’s t test with Welch’s correction. c PP1A activity was determined by using the specific assay. d Cardiomyocytes were co-transfected with LncDACH1 siRNA and PP1A siRNA for 48 h. PP1A siRNA successfully abrogated the influence of LncDACH1 siRNA on the proliferation of cultured cardiomyocytes by pH3 staining which is the marker of mitosis. n = 3 per group, one-way ANOVA followed by Newman–Keuls’s Multiple Comparison tests. e Overexpression of LncDACH1 (n = 5) increases the expression levels of p-YAP1, and silencing of LncDACH1 (n = 15) reduces the expression levels of p-YAP1. Two-tailed Student’s t test for parameter analysis, Mann–Whitney two-tailed U test for nonparametric analysis. f Calyculin A affected YAP1’s subcellular localization. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars = 50 μm.
To further investigate whether PP1A mediates the regulatory role of LncDACH1 in cardiac regeneration, we co-transfected LncDACH1 siRNA and PP1A siRNA in cardiomyocytes. The results showed that PP1A siRNA canceled the pro-proliferative effects of LncDACH1 silencing, as manifested by reduced pH3 positive cells (Fig. 7d). To further verify PP1A’s function, the pharmacological inhibitors of protein phosphatase 1, calyculin A, and okadaic acid were used to block PP1A activity. As shown in Supplementary Fig. 9, both calyculin A and okadaic acid inhibited the pro-proliferative effects of LncDACH1 silencing. These data indicate that PP1A is involved in the regulation of cardiomyocyte proliferation induced by LncDACH1 siRNA.
LncDACH1 regulates YAP1 phosphorylation by antagonizing PP1A
Hippo/YAP1 pathway has been shown to play a critical role in cardiac regeneration [6], and protein phosphatase is involved in regulating the dephosphorylation of Hippo signal pathway [25]. Thus, we further investigated if LncDACH1 binds PP1A and affects the dephosphorylation of YAP1. Our results showed that overexpression of LncDACH1 increased the phosphorylation of YAP1 protein, while produced no effect on total YAP1 protein level (Fig. 7e). The data suggest that LncDACH1 overexpression reduces its nucleus distribution and inactivates Hippo/YAP1 signal pathway. In contrast, knockdown of LncDACH1 increased the nucleus distribution of YAP1 protein in cardiomyocytes, which was abrogated by protein phosphatase 1 inhibitor calyculin A (Fig. 7e, f). These results indicate that LncDACH1 induces YAP1 phosphorylation by binding PP1A. Then, we further evaluated the involvement of YAP1 in LncDACH1 deficiency mediated cardiac regeneration in cultured neonatal cardiomyocytes. Inhibition of YAP1 by verteporfin reduced LncDACH1 siRNA mediated cardiomyocyte proliferation, as indicated by reduced pH3 positive and increased binucleated cells (Supplementary Fig. 10). We also analyzed the expression of YAP1’s target genes Ctgf, Birc5, Cyr61, Map5, and Ankrd1. LncDACH1 overexpression decreased the level of Ctgf, and silencing of LncDACH1 increased the expression level of cyr61 and ankrd1 (Supplementary Fig. 11). These results imply that LncDACH1 deficiency-induced cardiomyocyte proliferation and cardiac repair mediated by YAP1 signaling was regulated by PP1A.
Molecular basis for LncDACH1 inhibition of PP1A activity in cardiomyocytes
To further characterize the molecular basis of LncDACH1-PP1A interaction in cardiomyocytes, we aimed to dissect out the functional region of LncDACH1 sequence that accounts for the effects of LncDACH1 on cardiac regeneration. Different fragments of LncDACH1 (LncDACH1-a to f) were constructed as indicated in Fig. 8a. Except LncDACH1-d, the other five LncDACH1 fragments inhibited the proliferation of cultured neonatal mouse cardiomyocytes as manifested by reduced number of pH3 and EdU positive cells (Fig. 8b). The pull-down experiment showed that five LncDACH1 fragments a, b, c, e, and f interacted with PP1A, while LncDACH1-d did not (Fig. 8c). These data indicate that sequence 835–1251 is the functional domain of LncDACH1. Interestingly, catRAPID graphic also predicted the interaction of LncDACH1 fragments f with PP1A (Fig. 8d). The catRAPID fragments (an algorithm based on individual interaction propensities of polypeptide and nucleotide sequence fragments) analysis revealed that the 978–1068 nucleotide positions of the LncDACH1 sequence may bind to the 205–256 amino acid residues of the PP1A protein with high propensities (Fig. 8e, Supplementary Fig. 12). Thus, we further examined the proliferative effect of LncDACH1-PP1A with binding site mutation (LncDACH1(835–1251) mut) on cardiomyocytes. The results showed that overexpression of LncDACH1(835–1251) reduced the percentage of pH3 positive cardiomyocytes, however, LncDACH1(835–1251) mut lost the inhibitory effect on cardiomyocyte proliferation (Fig. 8f). These results indicate that the 978–1068 nucleotide position of the LncDACH1 sequence is the functional interaction sites of LncDACH1 and PP1A in hearts.
a LncDACH1 fragments (LncDACH1-a–f) were constructed. Full-length LncDACH1 is divided into five fragments. Fragment b, 418–2085 nt; fragment c, 835–2085 nt; fragment d, 1251–2085 nt; fragment e, 835–1668 nt; fragment f, 835–1251 nt. b The influence of LncDACH1 fragments on neonatal cardiomyocyte proliferation. Cardiomyocytes were transfected with different fragments and stained with pH3 for mitosis and EdU for DNA synthesis. One-way ANOVA followed by Dunnett’s post hoc test for multiple comparison. c Representative western blot analysis of RNA streptavidin pull-down assay. LncDACH1 fragments a–f were labeled with biotin followed by streptavidin bead pulldown. PP1A was detected by immunoblotting in the pulldown products. d Prediction of RNA-protein interaction of LncDACH1 with PP1A using the catRAPID graphic. e CatRAPID fragment module prediction of the interaction profile between PP1A protein and LncDACH1. f Mutation of LncDACH1 and PP1A binding site (LncDACH1(835–1251) mut) produced no effect on neonatal cardiomyocyte proliferation by pH3 staining. Data are shown as mean ± SEM, n = 5/group. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA followed by Newman–Keuls’s Multiple Comparison tests. Scale bars = 50 μm.
Effects of conserved sequence of LncDACH1 on human cardiomyocyte proliferation
The functional fragment of LncDACH1 (835–1251) also shows the higher conservation on the sequence between human and mice (Supplementary Fig. 13). We thus evaluated the effects of functional conserved sequences of LncDACH1 in the human genome (hLncDACH1) on the proliferation of human cardiomyocytes derived from induced pluripotent stem cells (iPS). As shown in Supplementary Figs. 14 and 15a,b, overexpression of hLncDACH1 reduced the percentage of both pH3- and EdU- positive cells, whereas the inhibition of hLncDACH1 promoted hiPS-CM proliferation. It suggests that the functional conserved fragment of LncDACH1 also causes the suppressive impact on human cardiomyocytes, which implies the possibility of targeting LncDACH1 in the treatment of ischemic heart diseases. In addition, we examined if the expression level of LncDACH1 in the plasma of patients is correlated with MI, and the results showed that the level of LncDACH1 in the plasma was higher in patients with MI than healthy groups (Supplementary Fig. 15c).
Discussion
In this study, we first revealed that the upregulation of LncDACH1 in postnatal and infarcted hearts suppressed cardiomyocyte mitosis and cardiac regeneration, while targeting LncDACH1 induces cardiomyocyte proliferation and heart regeneration after injury. LncDACH1 directly binds PP1A to influence the dephosphorylation and nuclear localization of YAP1, thereby inhibiting cardiomyocyte proliferation. The sequence fragment of 835–1251 nt of LncDACH1 is the key functional domain that interacts with PP1A. The functional conserved fragment of LncDACH1 also inhibits the proliferation of human iPS-derived cardiomyocytes. These findings extend our understanding of the molecular mechanisms for the loss of cardiac regenerative capacity in postnatal hearts, and provide a new therapeutic target for ischemic heart diseases.
The reactivation of cardiomyocyte proliferation is the critical mechanism responsible for inducing neonatal and adult heart regeneration. Meis1, Pitx2, Tbx20, and GATA-4 have been shown to play a pivotal role in regulating the proliferative capacity of mammalian cardiomyocytes [2, 4,5,6,7]. MiRNAs also play a vital role in the regulation of cardiomyocyte cell cycle and heart regeneration [8, 9, 26]. Moreover, several studies have demonstrated that lncRNAs participate in cardiac regeneration. In this study, we found that heart failure related LncDACH1 in our recent report [24] was obviously upregulated during postnatal development in mouse hearts and regulated cardiac regeneration. Our findings suggest that targeting LncDACH1 might be a novel strategy for inducing cardiac regeneration and reducing infarct size in mouse hearts. Of course, whether the improved heart function is related to the effect of LncDACH1 on cell death or other mechanisms is still unclear. More detail mechanisms will be studied in our future work.
Numerous studies have revealed that lncRNAs regulate biological activities and cellular functions by multiple mechanisms. For example, lncRNAs can act as competitive endogenous RNAs (ceRNA), regulate adjacent genes in cis, recruit chromatin modifier, inhibit translation, and modulate splicing [14, 16]. We recently identified lncRNA-CAREL as a pivotal molecule to regulate cardiac regeneration by binding miR-296 [27]. However, besides the ceRNA mechanism, the other new mechanisms underlying lncRNAs’ regulation of myocardial regeneration remain to be elucidated. Here, we found that LncDACH1 acts as a scaffold to bind PP1A, and suppresses the proliferation of cardiomyocytes. Consistently, PP1A as a certain class of protein serine/threonine phosphatases are shown to play an essential role in the control of muscle contraction, cell differentiation, cell cycle, apoptosis, and the regulation of membrane channels [28]. Particularly, PP1A’s sequence is highly conserved among eukaryotes. PP1A can form complexes with other regulatory subunits, indicating a critical role of PP1A in mediating cellular physiology and pathology. Consistently, pharmacological inhibition of PP1A or PP1A siRNA abrogates the increased mitosis of cardiomyocyte after LncDACH1 silencing, suggesting that LncDACH1 binds PP1A and inhibits its catalytic activity in cardiac myocytes.
The Hippo/YAP1 pathway plays a critical role in development, growth, repair, and homeostasis of tissues and organs [29]. As a core transcriptional regulator, YAP1 is involved in cardiac biological and pathological processes such as differentiation, proliferation, and apoptosis by activating the transcription of genes [29, 30]. Interestingly, we found that LncDACH1 overexpression upregulated p-YAP1, and YAP1 inhibitor verteporfin blocked the increased cardiomyocyte proliferation induced by LncDACH1 siRNA. Our results indicate that YAP1 acts as the downstream factor of LncDACH1-PP1A interaction, and is involved in cardiac regeneration loss due to LncDACH1 upregulation. This is the first study to reveal LncDACH1 as a PP1A inhibitor to regulate myocardial regeneration by inducing the cytoplasmic retention of YAP1. This study also firstly established the correlation of lncRNAs with YAP1 signaling in heart diseases. The plasma level of lncRNAs has emerged as one novel biomarker for ischemic heart diseases. We also found that the plasma level of LncDACH1 is remarkably increased during MI of mice and patients (Supplementary Figs. 3 and 15c), suggesting a close correlation between cardiac injury and LncDACH1 expression.
The clinical application of lncRNAs is normally limited by the lack of sequence conservation among human and animal species. In this study, we found the sequence of the functional domain of mouse LncDACH1 is highly conserved between human and mice. In consistence with the role of full-length LncDACH1 in mouse cardiomyocytes, the functional conserved fragment of LncDACH1 in human genome (hLncDACH1) also plays an inhibitory role in the proliferation of human cardiomyocytes derived from iPS cell lines. Likewise, it has been previously reported that the partially conserved sequence of human lncRNAs holds the same function as the full-length mouse lncRNAs [23, 27]. Our findings imply the potential of clinical application of targeting LncDACH1 to induce cardiac regeneration after MI.
There are still some limitations in our study. The application of α-actinin alone is one limitation of our research to identify the proliferative cardiomyocytes, although a large number of studies used α-actinin to label cardiomyocytes [11, 26, 30]. PCM-1, a marker of CM nuclei, has been used to label cardiomyocytes in recent studies [31, 32], while the use of PCM-1 alone is also not accurate enough to count proliferative cardiomyocyte. The combination of α-actinin and PCM1 to label cardiomyocytes will be used to overcome this limitation in our future study. Moreover, the method that distinguishes cardiomyocyte cytokinesis from mitosis we used is Aurora B staining, which appears throughout the process of cell division and acts as the marker of cytokinesis only when it located between the two daughter cells. Considering Aurora B’s limitation in assessing cytokinesis, other methods to better mark the cytokinesis of cardiomyocytes are still lacking in this field and remain to be developed.
In summary, our study uncovers that targeting LncDACH1 in postnatal and infarcted hearts induces a remarkable increase in PP1A activities and thereby YAP1 function, leading to reactivation of cardiomyocyte proliferation and heart regeneration. These findings provide a novel insight into the functional roles of lncRNAs in myocardial repair and regeneration, and suggest lncRNAs as potential targets for heart regeneration after injury.
References
Heusch G, Libby P, Gersh B, Yellon D, Bohm M, Lopaschuk G, et al. Cardiovascular remodelling in coronary artery disease and heart failure. Lancet. 2014;383:1933–43.
Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–53.
Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80.
Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534:119–23.
Lu F, Langenbacher A, Chen JN. Tbx20 drives cardiac progenitor formation and cardiomyocyte proliferation in zebrafish. Dev Biol. 2017;421:139–48.
Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7:279ra38.
Yu W, Huang X, Tian X, Zhang H, He L, Wang Y, et al. GATA4 regulates Fgf16 to promote heart repair after injury. Development. 2016;143:936–49.
Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest. 2013;123:1939–51.
Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–85.
Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, et al. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res. 2015;117:450–9.
Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66.
Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–79.
Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–7.
Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–89.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–81.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69.
van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–28.
Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83.
Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. 2015;89:98–112.
Marketou ME, Kochiadakis GE, Maragkoudakis S, Konstantinou J, Vardas PE, et al. The long non-coding RNAs MHRT, FENDRR and CARMEN, their expression levels in peripheral blood mononuclear cells in patients with essential hypertension and their relation to heart hypertrophy. Clin Exp Pharm Physiol. 2018;45:1213–7.
Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14.
Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121:575–83.
Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–9.
Cai B, Zhang Y, Zhao Y, Wang J, Li T, Zhang Y, et al. Long noncoding RNA-DACH1 (Dachshund Homolog 1) regulates cardiac function by inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a). Hypertension. 2019;74:833–42.
Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013;6:rs15.
Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–81.
Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, et al. The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. 2018;72:534–50.
Liu CY, Lv X, Li T, Xu Y, Zhou X, Zhao S, et al. PP1 cooperates with ASPP2 to dephosphorylate and activate TAZ. J Biol Chem. 2011;286:5558–66.
Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110:13839–44.
Li J, Gao E, Vite A, Yi R, Gomez L, Goossens S, et al. Alpha-catenins control cardiomyocyte proliferation by regulating Yap activity. Circ Res. 2015;116:70–9.
Sampaio-Pinto V, Rodrigues SC, Laundos TL, Silva ED, Vasques-Novoa F, Silva AC, et al. Neonatal apex resection triggers cardiomyocyte proliferation, neovascularization and functional recovery despite local fibrosis. Stem Cell Rep. 2018;10:860–74.
Zacchigna S, Martinelli V, Moimas S, Colliva A, Anzini M, Nordio A, et al. Paracrine effect of regulatory T cells promotes cardiomyocyte proliferation during pregnancy and after myocardial infarction. Nat Commun. 2018;9:2432.
Acknowledgements
This work was supported by the National Key R&D Program of China [2017YFC1307403, 2017YFC1307404], the National Natural Science Fund of China [81872857, 81573434, 81870295, and 81730012] and China Postdoctoral Science Foundation [2018M641867].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by H.-U. Simon
Supplementary information
Rights and permissions
About this article
Cite this article
Cai, B., Ma, W., Wang, X. et al. Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death Differ 27, 2158–2175 (2020). https://doi.org/10.1038/s41418-020-0492-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-0492-5
This article is cited by
-
N-Acetyltransferase 10 represses Uqcr11 and Uqcrb independently of ac4C modification to promote heart regeneration
Nature Communications (2024)
-
Cyclin L1 controls cardiomyocyte proliferation and heart repair after injury
Signal Transduction and Targeted Therapy (2023)
-
The role and medical prospects of long non-coding RNAs in cardiovascular disease
Heart Failure Reviews (2023)
-
Long noncoding RNA MALAT1 polymorphism predicts MACCEs in patients with myocardial infarction
BMC Cardiovascular Disorders (2022)
-
Cytoplasmic sequestration of p53 by lncRNA-CIRPILalleviates myocardial ischemia/reperfusion injury
Communications Biology (2022)