Abstract
Notch, an essential factor in tissue development and homoeostasis, has been reported to play an oncogenic function in a variety of cancers. Here, we report ubiquitin-specific protease 8 (USP8) as a novel deubiquitylase of Notch1 intracellular domain (NICD). USP8 specifically stabilizes and deubiquitylates NICD through a direct interaction. The inhibition of USP8 downregulated the Notch signalling pathway via NICD destabilization, resulting in the retardation of cellular growth, wound closure, and colony forming ability of breast cancer cell lines. These phenomena were restored by the reconstitution of NICD or USP8, supporting the direct interaction between these two proteins. The expression levels of NICD and USP8 proteins were positively correlated in patients with advanced breast cancer. Taken together, our results suggest that USP8 functions as a positive regulator of Notch signalling, offering a therapeutic target for breast cancer.
Similar content being viewed by others
Introduction
Notch signalling, a highly conserved pathway in evolution, plays an important role in the regulation of systemic homoeostasis, development, and proliferation in most tissues [1,2,3,4]. The Notch gene was first described in 1917 by Thomas Morgan, as partial loss of function resulting from notches at the end of the wing blades in a strain of Drosophila melanogaster [5]. The Notch gene encodes a transmembrane (TM) receptor with a molecular weight of 300 kDa. The core Notch signalling pathway is activated by cell-to-cell interaction, by which a ligand on one cell induces a series of proteolytic cleavage events in the cell containing a Notch receptor [6]. Through sequential cleavages from S1 to S3, the Notch intracellular domain, NICD, is released into the cytoplasm and translocates to the nucleus to activate the transcription of Notch target genes in association with CBF1/RBP-J, Su(H), Lag-1 (CSL) and Mastermind-like protein [7,8,9]. The activation of Notch signalling induces hairy/enhancer of split (HES) gene expression leading to systemic development and proliferation [8,9,10,11]. Thus, it is expected that modulating the stability of NICD is a potential way to regulate the Notch pathway. In addition to its effects on development, Notch is also well known for its oncogenic function. The elevated expression of NICD is closely related to poor prognosis in various types of cancer, including tumours of the breast, lung, and colon [12,13,14,15]. Therapeutic approaches targeting Notch for the inhibition of its function are currently being greatly developed [16]. Most clinical trials are currently evaluating Notch-targeting therapies using either γ-secretase inhibitor (GSI) or α-secretase inhibitor, which block the cleavage pathway of Notch to NICD [17,18,19,20]. Ubiquitylation and deubiquitylation are highly coordinated processes, which allow for ubiquitin to be recycled. The substrates regulated via these processes are subject to a variety of fates, including stabilization, degradation, or inhibition [21,22,23]. As Notch protein degradation occurs mostly through ubiquitylation, E3 ligases of Notch may play an important role in determining the rate of turnover of the Notch signalling system [24]. For example, F-box and WD repeat domain-containing 7 (FBXW7;FBW7) is a well-known E3 ligase for NICD [25, 26]. FBW7 has been reported to directly interact with NICD, promoting its proteasomal degradation and poly-ubiquitylation status [27, 28]. In addition, cytoplasmic E3 ligases such as Itch and Deltex also ubiquitylate Notch, which is mostly present in cell membranes or intracellular endocytic vesicles [29, 30]. Meanwhile, little is known about the role of deubiquitylating enzymes (DUBs) in the modulation of NICD. DUBs such as eIF3f and USP12 have been known to regulate Notch signalling through mono-ubiquitylation without any implication in protein stabilization [31, 32]. Markus et al. reported that USP28 is a deubiquitylase of c-Myc together with c-Jun and NICD in murine and human intestinal cancer [33]. However, its biochemical effect on NICD and clinical implications in cancer have not been fully explored.Ubiquitin-specific protease 8 (USP8), also known as UBPY, is a deubiquitylating enzyme that is involved in the growth regulation of various cancer cells [34]. Emi Mizno et al. reported that USP8 negatively regulates the ubiquitylation and lysosomal degradation of epidermal growth factor receptor (EGFR) [35]. USP8 knockout mice show embryonic lethality due to significant growth retardation and reduction of RTKs, such as EGFR, ERBB2, ERBB3, and c-MET [36]. USP8 is also known to be involved in modulating extrinsic apoptosis by enhancing Flip stability [37]. Recent studies have suggested that the depletion of USP8 leads to the suppression of STAM, HRS, RNF41, and Parkin, which leads to general cell growth retardation and cell death [36, 38,39,40]. Elevated expression of USP8 correlates with a poor prognosis in various cancer cells, suggesting that USP8 may be oncogenic and a potential target protein for cancer therapy [41, 42].In this study, we identified USP8 as a direct regulator of NICD. Loss of USP8 in breast cancer cell lines decreased cell growth, colony forming ability, and wound closure, which were restored by NICD reconstitution. Moreover, a combination of high USP8 and high NICD expression is associated with poor outcomes in breast cancer patients. Supporting these findings, USP8 was capable of stabilizing NICD by enhancing cellular proliferation. Collectively, these data suggest that USP8 functions as a positive regulator of NICD, offering a potential therapeutic opportunity for the treatment of cancer.
Materials and methods
Plasmids
A pCMV-FLAG-USP8 WT construct was kindly provided by KyungAh Kim (Samsung Advanced Institute of Technology, South Korea). pCMV-MYC-NICD constructs were generously provided by Guhung Jung (Seoul National University, South Korea). A FLAG-USP8 C786S mutant was generated via site-directed mutagenesis according to the manufacturer’s instructions (Takara Bio, Shiga, Japan). A MYC-tagged NICD WT from the pCMV-MYC vector was also subcloned into the pBABEpuro vector. pHM6-HA-Ub has been described previously [43]. siRNA-resistant-USP8#1 and USP8#3 variants from pCDH vector were generated via site-directed mutagenesis. pEGFP-C2 (Clontech, San Diego, CA, USA) was used as a transfection control.
Antibodies and chemicals
The following antibodies were used for western blot and immunoprecipitation assays: Cleaved-Notch1 (Val1744) (4147s; Cell Signaling, Danvers, MA, USA; 1:1,000), Notch1 (C-20) (sc-6014; Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:500), Notch1 (D1E11) (3608 s; Cell Signaling, 1:1,000), USP8 (A302-929A; Bethyl Laboratories, Montgomery, TX, USA; 1:2,000), HES1 (H-160) (sc-25392; Santa Cruz; 1:500), EGFR (4267; Cell Signaling; 1:1,000), phospho-EGFR (Tyr1068) (3777; Cell Signaling; 1:1,000), GAPDH (14C10) (2118; Cell Signaling, 1:5,000), Larmin A/C (2032; Cell Signaling, 1:2,000), MYC (sc-40; Santa Cruz, 1:1,000), HA (sc-805 rabbit; Santa Cruz, 1:1,000), FLAG (F3165 mouse; Sigma, 1:5,000, F7425 rabbit; Sigma, 1:2,000, A8592 HRP-conjugated; 1:2,000; Sigma-Aldrich, St. Louis, MO, USA), GFP (sc-8334; Santa Cruz; 1:1,000), β-actin (A5316; Sigma, 1:5,000), and normal rabbit IgG (sc-2027; Santa Cruz). MG132 and gefitinib were purchased from A.G. Scientific (San Diego, CA, USA), cycloheximide (CHX; C4859), N-ethylmaleimide (NEM; E3876), and dimethyl sulfoxide (DMSO; D8418) were purchased from Sigma-Aldrich. DAPT (GSI-IX) was purchased from Selleck Chemicals (S2215; Houston, TX, USA), and USP8 inhibitor (DUB-IN-2) was purchased from MedChem Express (Monmouth Junction, NJ, USA).
Cell culture and transfection
The H1299 (human nonsmall cell lung carcinoma) and the 293T (embryonic kidney cell line) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). The H1299 cells were cultured in the Roswell Park Memorial Institute (RPMI) medium (GIBCO) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS) (Thermo Scientific, Grand Island, NY, USA). The 293T cells were maintained in Dulbecco’s modified Eagle’s medium (Corning Cellgro, Manassas, VA, USA) supplemented with 10% FBS and 1% PS. The MDA-MB-231 (breast cancer cell line) was obtained from the US National Cancer Institute (NCI; MTA no. 2702-09) and was cultured in RPMI supplemented with 5% FBS and 1% PS. All cell lines were maintained in 5% CO2 at 37 °C and were treated with plasmosin (Invitrogen, Carlsbad, CA, USA) to prevent infection of mycoplasma. Polyethylenimine (Sigma-Aldrich) was used for 293 T transfection, including retrovirus and lentivirus production. Lipofectamine 2000 (Invitrogen) was used to transfect H1299 cells. siRNA was transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol.
siRNAs and shRNAs
USP8 siRNA #1 (5′-CAGGGTCAATTCAAATCTACA-3′), #3 (5′-GGTTCAGGCAAGCCATTTA-3′), and control siRNA were synthesized by Qiagen (Valencia, CA, USA). USP8 shRNA #1 (5′-TRCN0000007435, GCTGTGTTACTAGCACTATAT-3′), #5 (5′-TRCN0000284769, GCTGTGTTACTAGCACTATAT-3′), and control shGFP were purchased from Sigma-Aldrich. Lipofectamine RNAiMax was used for siRNA transfection according to the manufacturer’s protocol (Invitrogen). Stable WT NICD-expressing cell lines were developed using retroviral vectors (pBabe-puro-NICD). The 293T cells were transfected with pBabe-puro together with Gag-pol and VSV-G-expressing vectors for 48 h to produce packaging retroviruses. pLKO.1 shRNA-expressing vector and packaging vectors were co-transfected into 293T cells for 48 h. Supernatants were harvested, filtrated, and co-transferred to MDA-MD-231 cells. To select for retrovirus-and lentivirus-containing cells, cell lines were incubated following 1 μg/ml puromycin (Sigma-Aldrich).
Quantitative reverse transcription-PCR analysis
RNA from H1299 and MDA-MB-231 cells were extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s guidelines. cDNA was amplified using 1 µg of total RNA using PrimeScript Reverse Transcriptase (2680A; Takara Bio), and analyzed using QuantiTech SYBR Green PCR Kit (204143; Qiagen) and real-time PCR (Rotor-GeneQ 2plex, Qiagen). Total RNA samples were analyzed for NOTCH1, USP8, HES1, p21, c-MYC, and GAPDH expression. The following primers (IDT, Coralville, IA) were used for qRT-PCR: NOTCH1, 5′-GTTGATCTCGCAGTTGGGT-3′ (forward), 5′-CAACATCAACGAGTGCTCCA-3′ (reverse), USP8, 5′-CCAAACTGAAGCGCTCCTAC-3′ (forward), 5′-CTGGTCCAGAACCTCCAAAA-3′ (reverse), HES1, 5′-TCACCTCGTTCATGCACTC-3′ (forward), 5′-GAAATGACAGTGAAGCACCTC-3′ (reverse), p21(CDKN1B), 5′-CGCACGTTTGACATCTTTCTC-3′ (forward), 5′-CTGTGTCTTTTGGCTCCGA-3′ (reverse), c-MYC, 5′-TCTTCCTCATCTTCTTGTTCCTC-3′ (forward), 5′- TCCTCGGATTCTCTGCTCTC-3′ (reverse), GAPDH, 5′-GATCATCAGCAATGCCTCCT-3′ (forward), 5′-TGTGGTCATGAGTCCTTCCA-3′ (reverse). All mRNA levels were normalized to those of GAPDH.
Cell fractionation
Cytoplasmic and nuclear fraction was conducted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (78835; Thermo Scientific) according to the manufacturer’s protocol.
Immunoprecipitation and ubiquitylation assays
For immunoprecipitation assays, cells were lysed with a lysis buffer composed of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 0.5% NP-40, 0.1% Na-deoxycholate, 1 mM EDTA, and protease inhibitors (2 µM leupeptin, 1 µM pepstatin A, 2 µM aprotinin, 200 µM PMSF). Cell lysates were immunoprecipitated with 1 µg of antibodies overnight at 4 °C and incubated with protein G Sepharose beads (GE Healthcare, Buckinghamshire, UK) for 2 h at 4 °C. The beads were centrifuged and eluted with 2× sample buffer then boiled for 10 min. Ubiquitylation assays with HA-Ub and endogenous ubiquitylation assays were performed under denaturation conditions. Using PBS containing NEM, cells were lysed by boiling in 1% sodium dodecyl sulfate (SDS) for 10 min. Lysates were produced by adding the protease inhibitors and lysis buffer described above.
Luciferase reporter assay
The luciferase assay was conducted using a luciferase assay kit (Promega, Madison, WI, USA). The luciferase reporter plasmids were under the control of 4XCSL-Luc (a four-time repeating section of the RBP-Jk target sequence, CGTGGGAA, with the luciferase gene), Hes1-Luc (–467 to +46 of the Hes-1 promoter with the luciferase gene).
Crystal violet staining, wound healing, colony forming assays
For crystal violet staining, cells were plated into six-well culture dishes and fixed in 1% glutaraldehyde (G6257, Sigma-Aldrich) for 10 min. After washing with phosphate buffered saline (PBS), cells were stained with 0.5% crystal violet for 20 min at room temperature. For the scratch assay, cells were cultured to confluence (80%) in six-well plates then scratched with a micropipette tip. The wound healing area was continuously monitored at 12 and 24 h using a microscope. Cells were grown at 37 °C under 5% CO2 conditions. For the colony forming assay, cells were seeded at 200 cells per well (six-well plates), incubated at 37 °C, 5% CO2 for 10–12 days. The medium was replaced with fresh medium every 3 days. Cells were stained with 0.5% crystal violet as described above.
Clinical samples
A total of 167 breast cancer cases were selected from the pathology case archive of Kangbuk Samsung Hospital based on the diagnosis and the quality of the available tissue on the paraffin blocks. This study was approved by the Institutional Review Board of Kangbuk Samsung Hospital (approval no. #2017-08-017; Seoul, South Korea). All procedures were conducted in accordance with the Declaration of Helsinki. The tumours were graded according to the modified Bloom-Richardson grading system [44]. The patients’ mean age at operation was 52.0 years old (range; 28–92 years) and the mean follow-up period after operation was 59.4 months (range; 2–89 months). The pathologic tumour grade was 1, 2, and 3 in 77 (46.1%), 82 (49.1), and 8 (4.8%) patients, respectively. The regional lymph node metastasis was reported in 86 (51.5%) patients. Fifteen (9.0%) patients underwent a distant metastasis and the recurrence occurred in 25 (15.0%) patients (Supplementary Table S1).
Immunohistochemistry
Tissue microarrays (TMAs) were constructed from 167 formalin-fixed, paraffin-embedded (FFPE) tissue specimens. Briefly, 2-mm-diameter tissue cylinders were cored from the tumour areas of each FFPE tissue block and manually arrayed into recipient blocks. The TMA sections were cut to a thickness of 5 μm followed by deparaffinization through xylene and dehydration with graded ethanol. Heat-induced antigen retrieval was performed in an antigen retrieval buffer of pH 6.0 (Dako, Carpinteria, CA, USA). The endogenous peroxidase activity was quenched with 3% H2O2 in water for 10 min, and the TMA sections for NICD were incubated with a protein block (Dako) for another 20 min. USP8 (rabbit polyclonal, ab228572, Abcam, 1:400) and NICD (rabbit polyclonal, ab8925, Abcam, 1:250) antibodies were applied to individual slides. The primary antibodies were incubated for 30 min (NICD) or 1 h (USP8) at room temperature. Immunohistochemical staining was performed on a Dako Autostainer (Dako) using the EnVision + Dual Link System-HRP (Dako) according to the standard protocol. Antigen–antibody reaction was visualized with 3,3′-diaminobenzidine and lightly counterstained with haematoxylin. The percentage of stained cells was recorded for each intensity of staining (from 0 to 3) for each TMA core and the histoscore (range; 0–300) was obtained by multiplying the intensity and the percentage. The mean values of the histoscore were used as the cut-off value of high expression.
Statistical analysis
Statistical analyses were performed using PASW Statistics 18 for Windows (version 18.0; IBM SPSS Inc., Chicago, IL, USA) and the R statistical package (version 3.2.3). To evaluate the association between the expression of USP8 and NICD and the clinicopathologic parameters, crosstabs, and Pearson’s chi-squared (χ2) test were used as needed. Survival distributions were plotted using the Kaplan–Meier method, and the relationship between survival and each parameter was analyzed with the log-rank test. A Cox proportional hazards model was created to identify independent predictors of survival. Statistical significance was defined as a p-value lower than 0.05.
Results
Notch signalling is downregulated upon USP8 depletion
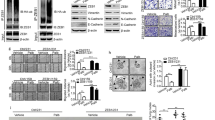
USP8 has been previously shown to enhance cell growth by stabilizing epidermal growth factor receptors [28]. The depletion of USP8 in cells grown under 10% FBS retarded MDA-MB-231 cell growth and destabilized EGFR and its phosphorylated form, as previously reported (Supplementary Fig. S1a–c) [35]. Interestingly, cell growth was further downregulated in the serum starved conditions with 0.1% FBS under which no EGFR was active, implicating that USP8 may have another target other than EGFR for enhancing cellular growth (Supplementary Fig. S1a–c). These observations led us to further investigate the role of USP8 independent of EGFR by employing gefitinib, an EGFR inhibitor, or via silencing of EGFR using EGFR siRNA. As shown in the Supplementary Fig. S1d–i, USP8 knockdown further restrained the cell growth under EGFR inhibition, confirming that USP8 may have other targets besides EGFR to support cell proliferation.To determine the molecular signalling pathways responsible for USP8 modulating cell growth in EGFR-independent manner, mRNA sequencing was conducted on MDA-MB-231 cells depleted of USP8 followed by gene set enrichment analysis (GSEA). Of a variety of pathways affected, the target genes associated with Notch signalling were shown to be prominently affected by the downregulation of USP8 (Fig. 1a). As previously reported, the signalling pathways associated with EGFR were also suppressed under the same conditions. Based on this result, we selected a variety of breast cancer cell lines, including MCF7, T47D, ZR-75-1, and SK-BR-3, and lung cancer cell lines, including A549, PC-9, H23, and H460, with physiological relevance to USP8 and NICD in different types of cancer. The data showed that, under the inhibition of USP8, all the cell lines displayed decreased protein levels of NICD (Supplementary Fig. S2a, c) and growth retardation (Supplementary Fig. S2b, d), indicating that the effect of USP8 on NICD could be plausible in both lung and breast cancer. These observations were further confirmed by transient or stable depletion of USP8 in either H1299 or MDA-MB-231 cells, a lung or breast cancer cell line, respectively. Corroborating with the RNA sequencing data, the protein levels of NICD as well as its target genes, including HES1, p21, and c-MYC, were downregulated upon USP8 inhibition in both cases (Fig. 1b and Supplementary Fig. S3a). On the other hand, the mRNA levels of Notch1 were not altered upon USP8 knockdown while the mRNA levels of other targets of Notch were reduced under USP8 depletion, indicating that USP8 may regulate NICD in the posttranslational step (Fig. 1c and Supplementary Fig. S3b). The knockdown of USP8 leading to the suppression of NICD transcriptional activities measured by the luciferase assay further confirmed that USP8 is associated with the Notch signalling pathway (Fig. 1d). The data suggest that the possibility of USP8 having effects on the protein levels of either Notch1 or its intracellular domain, or both. Is it worth noting that the protein levels of NICD were reduced under USP8 depletion, suggesting that USP8 could modulate only NICD protein (Fig. 1e). However, knockdown of USP8 did not affect full length Notch1 or Notch TM protein levels with or without DAPT-a novel GSI further proving that USP8 specifically targets NICD (Fig. 1e). Moreover, cytoplasmic and nucleus fractionation IP assay showed that USP8 interacted with NICD in the nucleus rather than in the cytoplasm (Fig. 1f). Lastly, USP8 depletion showed no effect on the surface levels of Notch1 (Supplementary Fig. S3c). Collectively, these data indicate that USP8 enhanced the stability of the NICD protein, which resulted in the enhancement of the Notch-dependent signalling pathways.
Depletion of USP8 regulates Notch signaling pathway. a Gene set enrichment analysis (GSEA) of NOTCH and EGFR target signatures using transcriptomes from MDA-MB-231 cells transfected with USP8 siRNA (20 nM). b Depletion of USP8 induces degradation of endogenous NICD protein levels. Stable knockdown of USP8 using shRNAs (shUSP8#1, shUSP8#5) in MDA-MB-231 cells also down-regulates endogenous NICD protein levels and indicated downstream targets genes. Each protein was detected by western blotting with the indicated antibodies. Relative NICD protein levels were quantified with ImageJ. c mRNA levels are shown in the right panel. Error bars indicate mean ± standard deviation (s.d.); n = 3, n.s = not significant, *p < 0.05, **p < 0.01 compared with shGFP. d Transient knockdown of USP8 downregulated Notch downstream promoter activities. After 24 h of gene silencing using control and USP8 siRNAs in H1299 cells, HES-1, HES-5, and 4×CSL plasmids were transfected. Cells were measured using a luminometer. P values were determined using a two-tailed Student t test (*p < 0.05, **p < 0.01). e The MDA-MB-231 cells transiently transfected with USP8 siRNA (siUSP8#1, siUSP8#3) and treated with 10 µM DAPT for 24 h were analyzed by western blotting. Cells were harvested at 72 h post transfection. f Cells were separated into cytoplasmic and nuclear proteins using fractionation kit and immunoprecipitated with indicated antibodies. MG132 was treated with 20 µM for 6 h before cell harvesting
USP8 functions as a deubiquitylase of NICD
The possibility of USP8 modulating NICD protein in the posttranslational stage led us to examine whether USP8 could interact with NICD. Both the exogenous and endogenous NICD and USP8 were demonstrated to interact by immunoprecipitation assays (Fig. 2a, b). NICD is composed of several domains, including two NLS domains. Based on this structural information, truncated forms of NICD mutants were constructed to identify the domain responsible for its interaction with USP8. The domain analyses showed that the Ankyrin domain of NICD was responsible for its interaction with USP8 (Fig. 2c, d). Next, we tested whether USP8 is capable of promoting the stabilization of NICD protein. The overexpression of USP8 led to the stabilization of exogenous (Fig. 3a) as well as endogenous protein levels of NICD (Fig. 3b), without any change of FL-Notch1 protein levels (Fig. 3b). These data were further confirmed by the treatment of cycloheximide (CHX), an inhibitor of the protein translation process. The half-life of overexpressed NICD and endogenous NICD protein was prolonged in the presence of USP8 while it was significantly diminished upon USP8 inhibition (Fig. 3c–e and Supplementary Fig. 4a, b). FL-Notch1 levels were not affected by the presence or absence of USP8 (Fig. 3c–e). Since USP8 is a deubiquitylase, next, we tested whether ubiquitin and proteasome-dependent pathways are involved in the interaction between USP8 and NICD. The observations that NICD being stabilized by MG132, the inhibitor of proteasome, was no longer being affected by the overexpressed USP8 indicated that there could be a ubiquitin and proteasome system involved in the USP8 and NICD interaction. Furthermore, the destabilized NICD upon USP8 depletion was restored by MG132 treatment, which also indicated that USP8 may regulate NICD levels, as suggested above (Fig. 3f, g). To further demonstrate the roles of USP8 in NICD regulation, wild-type (WT) USP8 and catalytically inactive mutant C786S (CS) were employed. As expected, only WT USP8 increased NICD protein levels (Fig. 3h) and its transcriptional activities (Fig. 3i), while the CS mutant had no effect. These events were irrelevant to its binding to NICD since both WT and its mutant were able to bind to NICD in a similar manner (Fig. 3j). These data suggest that the deubiquitylase activity of USP8 is required for stabilizing NICD. Next we tested whether USP8 could mediate the deubiquitylation of NICD. Poly-ubiquitylated forms of NICD accumulated upon MG132 treatment were almost diminished by USP8 regardless of the presence or absence of MG132 (Fig. 4a). These experiments were conducted under denaturation conditions, suggesting that there were no nonspecific interactions involved. As FBW7 is a well-known E3 ligase of NICD, we tested whether FBW7-mediated degradation of NICD could be rescued by the presence of USP8. The data showed that USP8 could restore the protein levels of NICD degraded by FBW7 and remove the ubiquitin chain from FBW7-mediated ubiquitylated NICD, suggesting that FBW7 and USP8 may be obligatory partners for the regulation of NICD (Fig. 4b, c). To test whether the protease activities of USP8 are involved WT USP8 and its protease defective mutant, the CS form was employed. The data shows that only the WT was able to mediate deubiquitylation of NICD, indicating that protease activities are directly required (Fig. 4d). In accordance with these data, only WT USP8, but not the CS mutant, decreased endogenous ubiquitylation status (Fig. 4e). Furthermore, USP8 depletion led to a greater accumulation of endogenous poly-ubiquitylated forms of NICD compared to the control (Fig. 4f, lanes 2 and 3). These processes were reserved by the overexpression of USP8 resistant to its own siRNA, indicating that USP8 is responsible for deubiquitylation (Fig. 4f, lane 4). In summary, these results demonstrate that USP8 functions as a bona fide deubiquitylase of NICD.
NICD interacts with USP8. a NICD and USP8 proteins interact with each other. FLAG-tagged USP8 alone or with MYC-tagged NICD plasmids were transfected into 293T cells. MG132 (20 µM) were treated for 8 h. Whole cell lysates were immunoprecipitated with FLAG- and MYC- antibody. b Endogenous USP8 and endogenous NICD bind to each other. The 293T cell lysates were immunoprecipitated with anti-NICD or anti-USP8. NICD WT and each deletion mutant plasmids were transfected into the 293T cells. c Diagram of NICD truncated mutant constructs. d Ankyrin region of NICD is required for interaction with USP8
USP8 is required for upregulation and stabilization of NICD. a NICD protein levels were increased in a USP8-dose dependent manner. The H1299 cells were transfected with mixtures of plasmids expressing MYC-tagged NICD and FLAG-tagged USP8 with increasing concentrations. 2× GFP was used as a transfection control. b USP8 induces endogenous NICD protein levels. The H1299 cells were transfected with FLAG-tagged USP8. Relative NICD protein levels were shown in the right panel. Error bars indicate mean ± s.d.; n = 3. c USP8 increases NICD protein stability. The H1299 cells were transfected with MYC-tagged NICD in the absence or presence of FLAG-tagged USP8 followed by 100 µg/ml cycloheximide (CHX) treatment before harvesting at the indicated time points. Cells were lysed and analyzed by western blotting. The relative amounts of NICD were analyzed using ImageJ, as shown in the right panel. Error bars indicate mean ± s.d.; n = 3. d Endogenous NICD protein stability was enhanced by USP8 overexpression. The H1299 cells were transfected with Mock or FLAG-tagged USP8 followed by 100 µg/ml CHX for the indicated times. The relative amounts of NICD are shown in the right panel. (e) Knockdown of USP8 decreases stability of endogenous NICD. The H1299 cells stably expressing shGFP and shUSP8#1 were treated with 100 µg/ml CHX treatment before harvesting at the indicated time points. Western blotting analysis was then performed using the indicated antibodies. The relative amounts of NICD are shown in the right panel. f Endogenous NICD protein levels were checked in the presence of FLAG-tagged USP8. The H1299 cells, transfected with the indicated plasmids, were treated with 20 µM MG132 for 8 h. Cells were lysed and measured using the indicated antibodies. g Knockdown of USP8 decreased endogenous NICD protein levels. This effect was rescued by MG132 (20 µM, 8 h). Lysed cells were immunoblotted. h Deubiquitylase activity of USP8 is required for the induction of endogenous NICD protein levels. Plasmids expressing FLAG-tagged USP8 WT or C786S mutant and MYC-tagged NICD were transfected into H1299 cells. i Transcriptional activities of HES-1 were measured by luciferase reporter analysis. The H1299 cells were transfected with indicated plasmids. P values were determined using a two-tailed Student t test (***p < 0.001). j Both USP8 WT and its catalytic mutant C786S interact with NICD. The 293T cells were transfected with mixtures of plasmids expressing MYC-tagged NICD, FLAG-tagged USP8 WT, or FLAG-tagged USP8 C786S, followed by immunoprecipitation using anti-FLAG antibodies. MG132 (20 µM) was treated for 8 h before cell harvesting. Immunoprecipitated proteins were detected with anti-MYC and anti-FLAG antibodies
NICD is deubiquitylated by USP8. a USP8 induces deubiquitylation of NICD. The H1299 cells were transfected with the indicated plasmids, treated with or without 20 µM MG132 for 8 h. Cell lysates were immunoprecipitated with anti-MYC antibodies, followed by western blotting analysis using HA-HRP, anti-MYC, or anti-FLAG antibodies. HA-tagged ubiquitination analysis was performed under denaturation conditions using 1% SDS buffer. b Protein levels downregulated by FBW7 were restored by USP8 overexpression. Cells were lysed and detected using the indicated antibodies. c FBW7-mediated ubiquitylation of NICD was deubiquitylated by USP8. Twenty micrometres MG132 was treated for 8 h. HA-tagged ubiquitination analysis was performed under denaturation conditions. d USP8 WT deubiquitylates NICD, whereas its mutant C786S does not. The H1299 cells were transfected with the indicated plasmids and treated with 20 µM MG132 for 8 h. Cell lysates were immunoprecipitated with anti-MYC antibodies, followed by western blotting analysis using HA-HRP, anti-MYC, or anti-FLAG antibodies. HA-tagged ubiquitination analysis was performed under denaturation conditions using 1% SDS buffer. e Endogenous NICD ubiquitylation upon FLAG-tagged USP8 WT or CS plasmids and treated with 20 µM MG132 for 8 h. Cells were immunoprecipitated with anti-NICD antibody, followed by western blotting analysis under denaturation conditions. f Depletion of USP8 induces ubiquitylation of NICD. Stable cell lines of the control and siRNA-resistant USP8 were used. Cells were transiently transfected as indicated with control and USP8#1 siRNAs for 48 h, then treated with 20 µM MG132 for 8 h
Cell growth retardation under USP8 ablation is restored by NICD
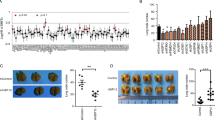
The biochemical data suggest that the overexpression of USP8 and NICD are closely correlated. Thus, we further analyzed the physiological relevance of these two factors using the MDA-MB-231 breast cancer cell line. First, MDA-MB-231 cell lines stably overexpressing pBabe control or WT NICD co-expressing with shUSP8#1, shUSP8#5 for stable knockdown of USP8 were generated (Fig. 5a). As expected, cell growth was downregulated in cells with USP8 depletion. However, this effect was rescued by NICD overexpression, even in USP8 knockdown conditions (Fig. 5a–c). As the protein levels of EGFR and phosphorylated EGFR were not changed when NICD was overexpressed (Fig. 5a), these regulatory effects are solely dependent on the presence of NICD. Similar results were obtained with the scratched wound healing and colony forming analyses indicating that the cellular proliferation retardation caused by USP8 ablation could be rescued by NICD overexpression (Fig. 5d, e). Taken together, these data strongly support the hypothesis that cancer cell proliferation is mediated by NICD stabilized by USP8.
USP8 regulates cell proliferation, colony forming ability, and wound healing ability. a The MDA-MB-231 cells were generated by pBabe-puro and pabe-NICD WT-expressing viral vectors using virus infection systems and stably expressing shGFP, shUSP8#1, and shUSP8#5 using retroviral vectors. Lysed cells were measured using the indicated antibodies. The data represent the average value from three independent experiments. b In total, 1 × 105 USP8-silenced MDA-MB-231 cells expressing NICD were plated equally, counted at each time point. Three independent experiments were performed in this setting. Error bars indicate the mean ± s.d.; n = 3. c Inhibition of USP8 led to the suppression of cell proliferation and showed growth recovery by NICD reconstitution. Cells were stained with 0.5% crystal violet. d Scratched wound healing assay upon USP8 silencing in pBabe or pBabe-NICD-expressing MDA-MB-231 cells. Wound closure was verified at each time point after the scratch. Dashed lines (white) represent the initial scratch wounds. Images were quantified by measuring intensity using ImageJ software (right panel). Data represent the mean ± s.d. of triplicates (**p < 0.01, ***p < 0.001). Scale bar, 100 µm. e Stably overexpressed NICD reversed a decrease in colony forming ability due to USP8 silencing. Cells were stained with crystal violet. Error bars indicate the mean ± s.d.; n = 3
Notch signalling pathway regulated by USP8 could be targeted to restrain cancer cell growth
As DUBs-IN-2 is a potent deubiquitylase inhibitor of USP8, next, we investigated whether DUBs-IN-2 could regulate NICD protein status. Endogenous NICD protein and its downstream target protein HES1 was dose-dependently degraded by DUBs-IN-2 treatment (Supplementary Fig. S5a). Accordingly, the endogenous ubiquitylated forms of NICD were significantly increased by DUBs-IN-2 (Supplementary Fig. S5b). We then investigated whether DUBs-IN-2 showed physiologically similar effects to USP8 depletion. DUBs-IN-2 downregulated NICD and HES1 protein levels (Supplementary Fig. S6a). Interestingly, the inhibitory effect of DUBs-IN-2 was mitigated by NICD overexpression. The inhibitory effects of DUBs-IN-2 treatment on cell growth, wound closure, and colony forming ability were rescued by NICD overexpression in the MDA-MB-231 cells (Supplementary Fig. S6b–e). These data indicate that the inhibition of USP8 by DUBs-IN-2 could suppress the stabilization of NICD and result in regression of cell proliferation. However, overexpression of NICD mitigated the inhibitory effect of DUBs-IN-2 on USP8 and stimulated cell growth and proliferation.To further analyze the positive effect of USP8 on NICD, MDA-MB-231 cells harbouring USP8 resistant to its own siRNA were made to exclude siRNA off-target effects. The protein levels of NICD and downstream target molecules were downregulated upon control cell line transfection with USP8 siRNAs, whereas cells expressing USP8 resistant to its own siRNA were unable to suppress their levels (Fig. 6a). Cell growth, scratched wound healing, and colony forming abilities were restored by the expression of USP8 in cells treated with USP8 siRNA (Fig. 6b–e). Corroborating these results, the overexpression of USP8 alone induced significant stabilization of NICD and HES1 proteins with the enhancement of cell growth (Supplementary Fig. S7). Collectively, these data suggest that USP8 exhibited an oncogenic effect by stabilizing NICD proteins, which resulted in the augmentation of its function leading to the proliferation of cancer cells.
USP8 positively regulates cell growth. a USP8 siRNA-resistant cell lines were obtained using a virus infection system in MDA-MB-231 cells. The data represent the average value from three independent experiments. b Cell growth curve data. Cells were counted at the indicated time points. Error bars indicate the mean ± s.d.; n = 3, **p < 0.01, ***p < 0.001. c Cells were stained with 0.5% crystal violet. d Scratched wound healing assay. Images were quantified by measuring intensity using ImageJ software (right panel). Data represent the mean ± s.d. of triplicates (***p < 0.001). Scale bar, 100 µm. e Colony forming ability. Cells were stained with crystal violet. Experiments were performed as described in Fig. 5b–e. The data represent the average value from three independent experiments. Error bars indicate the mean ± s.d.; n = 3, ***p < 0.001
USP8 is overexpressed in breast cancer patients
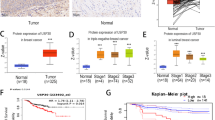
Based on these observations, we assessed the clinical significance of USP8 and NICD expression levels in the tissues of patients with breast cancer. We examined their expression levels in the TMA by immunohistochemistry (Fig. 7a), and analyzed the correlation between USP8 and NICD. Dual USP8 and NICD immunoreactivity was significantly correlated with distant metastasis (p = 0.005), estrogen receptor status (p = 0.039), and progesterone receptor status (p = 0.001). The expression level of USP8 exhibited a moderate positive correlation with that of NICD (Spearman’s ρ = 0.252, p = 0.002) (Fig. 7b). The 5-year recurrence-free survival (RFS) rate for patients exhibiting enhanced expression levels of USP8 (77.1%) and NICD (79.8%), was significantly lower than that for patients exhibiting low expression levels of USP8 (91.9%) and NICD (89.7%) (p = 0.018 and p = 0.070, Fig. 7c). Furthermore, patients exhibiting double positive expression had significantly shorter 5-year RFS rate (71.2%) when compared with that of patients exhibiting double negative (92.3%) and single positive (90.5%) (p = 0.004, Fig. 7c) expression. The Cox proportional hazard model demonstrated that the combination of USP8high and NICDhigh expression (hazard ratio = 1.651) (95% confidence interval (CI), 1.064–2.563, p = 0.025) is an independent predictor of shorter RFS (Supplementary Fig. S8). These data indicate that USP8 expression positively correlated with NICD expression and that USP8 potentially plays an important role in breast tumorigenesis.
Clinical significance of USP8-NICD expression in breast cancer patients. Protein expression levels of USP8 and NICD were determined using immunohistochemical analysis. a Representative immunohistochemical images stained with USP8 and NICD in breast cancer tissues. High magnification images are shown in inset. Scale bar, 100 µm. b The expression level of USP8 positively correlated with that of NICD (Spearman’s ρ = 0.252, p = 0.002). c Patients exhibiting USP8high (5-year RFS rate; 77.1%, p = 0.018) and USP8high/NICDhigh (5-year RFS rate; 71.2%, p = 0.004) expression had significantly shorter recurrence-free survival than patients exhibiting USP8low (5-year RFS rate; 91.9%), double negative (5-year RFS rate; 92.3%) and single positive (5-year RFS rate; 90.5%) expression
Discussion
Notch plays an essential function in various human cancers in a tissue- or cellular context-dependent manner [13, 45]. Studies have demonstrated the oncogenic roles of NICD in many types of cancer, including breast, lung, and colon cancer [46,47,48,49,50,51]. Because aberrant activation of Notch signalling in these cancers is associated with poor prognosis, identifying the factors and their associated mechanisms involved in the stabilization of Notch or NICD could provide potential therapeutic targets. The protein homoeostasis of NICD has been reported to be modulated by FBW7 [52, 53], a potent E3 ubiquitin ligase which also targets major oncogenes such as c-Myc, c-Jun, and Cyclin E [54,55,56,57,58,59]. Here, we found that USP8 restores the protein levels of NICD and suppresses ubiquitylated forms of NICD offsetting to action of FBW7, suggesting the obligatory partnership between FBW7 and USP8 (Fig. 4b, c). In addition to a previous report where USP8 was found to regulate cell growth through EGFR [36, 38,39,40], we demonstrated that USP8 regulates growth in an EGFR-independent manner (Supplementary Fig. S1). RNA-seq analyses revealed that this event was strongly related to the regulation of the Notch signalling pathway (Fig. 1a). Notch signalling was downregulated upon USP8 depletion, without any changes to FL-Notch1 protein, mRNA, or surface levels. These results indicate that USP8 specifically regulates NICD, and that this event is a unique and interesting phenomenon because the levels of FL-Notch1 and Notch-IC normally change together. Our study suggests that USP8 is a deubiquitylating enzyme of NICD by directly inducing stabilization and deubiquitylation through posttranslational modifications (Figs. 3 and 4). The physiological correlation between the USP8 and NICD signalling pathways is shown by the retardation of cellular growth, colony formation, and wound healing ability led by USP8 depletion or its inhibitor (DUBs-IN-2) and the subsequent rescue by the re-introduction of NICD (Fig. 5 and Supplementary Fig. S6).Cross-talk between Notch and EGFR in non-small cell lung cancer (NSCLC) has been reported by many research groups attempting to elucidate the association between the two different mechanisms in a tissue-dependent manner [60,61,62]. For example, EGFR activation may negatively regulate the Notch-dependent pathway or Notch could up-regulate the expression of EGFR, thereby enhancing tumour apoptosis or tumour cell growth, respectively [63]. The fact that both the Notch and EGFR signalling pathways may be regulated by USP8 indicate that both signalling pathways could be simultaneously enhanced by USP8 expression, possibly augmenting cellular proliferation. Indeed, a positive correlation between NICD and USP8 was determined in several NSCLC and breast cancer cells (Supplementary Fig. 2). In support of this data, the low survival rate of the breast cancer patients expressing both high levels of USP8, and combination of USP8 and NICD addresses the possible association and oncogenic roles both two proteins in human cancer (Fig. 7). The biochemical and physiological data indicate that USP8 may enhance the oncogenic intensity of NICD in combination with EGFR in various types of cancer, including breast cancer, and is a potential target for use in therapeutic trials.The fact that USP8 acts as a deubiquitylase for both NICD and EGFR is particularly interesting and noteworthy since the synergistic effects of both proteins have been reported in several types of cancer. Previously, it was reported that Notch1 regulates EGFR through p53 in gliomas, and that p53-null cells had no significant changes in EGFR mRNA levels or promoter activities. However, the fact that USP8 was found to enhance the protein stability of both proteins suggest the existence of alternative pathways to EGFR and NICD [64]. The p53-null cell lines, H1299 and MDA-MB-231, used in our experiments clearly show that either USP8 depletion or overexpression in these cells resulted in the destabilization or stabilization of both NICD and EGFR, respectively. Clinically, efforts have been taken to develop therapeutic strategies for various cancers employing a combination of treatment with both NICD inhibitors and EGFR inhibitors. For example, combination therapy such as γ-secretase inhibitor RO4929097 with erlotinib is currently a phase I/II clinical trial [65]. DAPT with gefitinib in NSCLC)has shown effective tumour growth retardation in vitro and in vivo [66]. Although the role of USP8 in enhancing the expression levels of NICD and EGFR would provide an alternative target to arrest the development of cancer in addition to the current combination trials, some issues must first be addressed in future studies. Firstly, the regulatory mechanism underlying the overexpression of USP8 in cancer, which was observed in our clinical data, must be elucidated. The clinical data in this study were analyzed using a relatively small sample size at a single institution. Thus, large-scale or multi-institutional studies including triple negative breast cancer samples are needed for validating the prognostic values of USP8 and NICD. Posttranslational as well as epigenetic changes must be evaluated to examine the role of various pathways. Secondly, clinical trials using DUBs-IN-2, an inhibitor of USP8 [67], must be established. The toxicity of the chemical could limit its usage at a systemic level. Furthermore, the suppression of USP8 may result in physiological defects, which would reduce the potential of USP8 as a good therapeutic target. To overcome this limitation, chemicals that regulate the protein interaction between USP8 and NICD or EGFR, rather than those that directly target USP8 catalytic activities, must be evaluated in future studies. Finally, in vivo studies using the USP8 conditional knockout mouse model or nude-mouse transplanted tumour model can aid in validating the oncogenic role of USP8 in various types of cancer.
References
Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17:722–35.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Raphael K, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33.
Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–51.
Morgan TH. The theory of the gene. Am Naturalist. 1917;51:513–44.
Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89.
Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–83.
Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;24:985–96.
Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71.
Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33.
Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–6.
Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7.
Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–35.
Al-Hussaini H, Subramanyam D, Reedijk M, Sridhar SS. Notch signaling pathway as a therapeutic target in breast cancer. Mol Cancer Ther. 2011;10:9–1.
Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–21.
Garber K. Notch emerges as new cancer drug target. J Natl Cancer Inst. 2007;99:1284–5.
Cook JJ, Wildsmith KR, Gilberto DB, Holahan MA, Kinney GG, Mathers PD, et al. Acute gamma-secretase inhibition of nonhuman primate CNS shifts amyloid precursor protein (APP) metabolism from amyloid-beta production to alternative APP fragments without amyloid-beta rebound. J Neurosci. 2010;30:6743–50.
Deangelo DJ, Stone RM, Silverman LB, Stock W, Attar EC, Fearen I et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J Clin Oncol. 2006; https://doi.org/10.1200/jco.2006.24.18_suppl.6585.
Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30:2307–13.
Schott AF, Landis MD, Dontu G, Griffith KA, Layman RM, Krop I, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin Cancer Res. 2013;19:1512–24.
Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–8.
Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol. 2015;16:322–4.
Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;8:579–86.
Moretti J, Brou C. Ubiquitinations in the notch signaling pathway. Int J Mol Sci. 2013;14:6359–81.
Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, et al. Functional interaction between SEL-10, an Fbox protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–8.
Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403–15.
Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel‑10 homolog. J Biol Chem. 2001;276:35847–53.
O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–24.
Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, et al. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–44.
Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–44.
Julien M, Patricia C, Stefano G, Sara FH, Annette MD, René Bernards, et al. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 2010;8:e1000545.
Moretti J, Chastagner P, Liang CC, Cohn MA, Israël A, Brou C. The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation. J Biol Chem. 2012;287:29429–41.
Diefenbacher ME, Popov N, Blake SM, Schülein-Völk C, Nye E, Spencer-Dene B, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Investig. 2014;124:3407–18.
Naviglio S, Mattecucci C, Matoskova B, Nagase T, Nomura N, Di Fiore PP, et al. UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 1998;17:3241–50.
Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–74.
Niendorf S, Oksche A, Kisser A, Lohler J, Prinz M, Schorle H, et al. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–39.
Jeong M, Lee E-W, Seong D, Seo J, Kim J-H, Grootjans S, et al. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIPL stability. Oncogene. 2017;36:458–70.
Ro PE, Prior IA, McCullough J, Clague MJ, Urbé S. The ubiquitin isopeptidase ubpy regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–24.
Wu X, Yen L, Irwin L, Sweeney C, Carraway KL. Stabilization of the e3 ubiquitin ligase nrdp1 by the deubiquitinating enzyme usp8. Mol Cell Biol. 2004;24:7748–57.
Durcan TM, Tang MY, Pérusse JR, Dashti EA, Aguileta MA, McLelland GL, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from Parkin. EMBO J. 2014;33:2473–91.
Byun S, Lee SY, Lee J, Jeong CH, Farrand L, Lim S, et al. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin Cancer Res. 2013;19:3894–904.
Yan Min, Zhao Cuihong, Wei Na, Wu Xiaoqian, Cui Jianli, Xing. Yanling. High expression of ubiquitin-specific protease 8 (USP8) is associated with poor prognosis in patients with cervical squamous cell carcinoma. Med Sci Monit. 2018;24:4934–43.
Yang MR, Lee SR, Oh W, Lee EW, Yeh JY, Nah JJ, et al. West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol. 2008;10:165–76.
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO classification of tumours of the breast, Volume 4. IARC: Lyon, France; 2012.
Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–9.
Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–59.
Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S et al. Alterations of the Notch pathway in lung cancer; Proc Natl Acad Sci. USA. 2009;106:22293–8.
Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676–85.
Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS, To KF, et al. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit Rev Oncol Hematol. 2013;86:251–77.
Zhang Y, Li B, Ji ZZ, Zheng PS. Notch1 regulates the growth of human colon cancers. Cancer. 2010;116:5207–18.
Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25.
Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–8.
Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–53.
Grim JE, Gustafson MP, Hirata RK, Hagar AC, Swanger J, Welcker M, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181:913–20.
Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25.
Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–8.
Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33.
Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7.
Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–22.
Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross‐talk. Crit Rev Biochem Mol Biol. 2006;41:339–85.
Konishi J, Yi F, Chen X, Vo H, Carbone DP, Dang TP. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene. 2010;29:589–96.
Kolev V, Mandinova A, Guinea‐Viniegra J, Hu B, Lefort K, Lambertini C, et al. EGFR signaling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–11.
Pancewicz-Wojtkiewicz J. Epidermal growth factor receptor and notch signaling in non-small-cell lung cancer. Cancer Med. 2016;5:3572–8.
Purow BW, Sundaresan TK, Burdick MJ, Kefas BA, Comeau LD, Hawkinson MP, et al. Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis. 2008;29:918–25.
Gold KA, Byers LA, Fan YH, Fujimoto J, Tse WH, Jack Lee J, et al. A phase I/II trial combining erlotinib with gamma secretase inhibitor RO4929097 in advanced non‐small cell lung cancer (NSCLC). J Clin Oncol. 2013;31 Suppl. 8104–8104.
Xie M, He CS, Wei SH, Zhang L. Notch‐1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non‐small cell lung cancer in vitro and in vivo. Eur J Cancer. 2013;49:3559–72.
Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, et al. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. Chem Med Chem. 2010;5:552–8.
Acknowledgements
This research was supported by grants from the Ministry of Science, ICT and Future Planning (NRF-2015R1A3A2066581) (JS). In addition, this research was partially supported by the BK21 Plus project of the National Research Foundation of Korea Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by P. Salomoni
Supplementary information
Rights and permissions
About this article
Cite this article
Shin, S., Kim, K., Kim, HR. et al. Deubiquitylation and stabilization of Notch1 intracellular domain by ubiquitin-specific protease 8 enhance tumorigenesis in breast cancer. Cell Death Differ 27, 1341–1354 (2020). https://doi.org/10.1038/s41418-019-0419-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0419-1
This article is cited by
-
FBP1 inhibits NSCLC stemness by promoting ubiquitination of Notch1 intracellular domain and accelerating degradation
Cellular and Molecular Life Sciences (2024)
-
USP8 positively regulates hepatocellular carcinoma tumorigenesis and confers ferroptosis resistance through β-catenin stabilization
Cell Death & Disease (2023)
-
Targeting ubiquitin-specific protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy of pancreatic cancer
Cell Death & Differentiation (2023)
-
USP8 inhibitor–induced DNA damage activates cell cycle arrest, apoptosis, and autophagy in esophageal squamous cell carcinoma
Cell Biology and Toxicology (2023)
-
USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy
Nature Communications (2022)