Abstract
Long noncoding RNAs (LncRNAs) have been implicated in the regulation of adipocyte and osteoblast differentiation. However, the functional contributions of LncRNAs to adipocyte or osteoblast differentiation remain largely unexplored. In the current study we have identified a novel LncRNA named peroxisome proliferator-activated receptor γ coactivator-1β-OT1 (PGC1β-OT1). The expression levels of PGC1β-OT1 were altered during adipogenic and osteogenic differentiation from progenitor cells. 5′- and 3′-rapid amplification of cDNA ends (RACE) revealed that PGC1β-OT1 is 1759 nt in full length. Overexpression of PGC1β-OT1 in progenitor cells inhibited adipogenic differentiation, whereas silencing of endogenous PGC1β-OT1 induced adipogenic differentiation. By contrast, overexpression of PGC1β-OT1 in progenitor cells stimulated, whereas silencing of PGC1β-OT1 inhibited osteogenic differentiation. In vivo experiment showed that silencing of endogenous PGC1β-OT1 in marrow stimulated fat accumulation and decreased osteoblast differentiation in mice. Mechanism investigations revealed that PGC1β-OT1 contains a functional miR-148a-3p binding site. Overexpression of the mutant PGC1β-OT1 with mutation at the binding site failed to regulate either adipogenic or osteogenic differentiation. In vivo crosslinking combined with affinity purification studies demonstrated that PGC1β-OT1 physically associated with miR-148a-3p through the functional miR-148a-3p binding site. Furthermore, PGC1β-OT1 affected the expression of endogenous miR-148a-3p and its target gene lysine-specific demethylase 6b (KDM6B). Supplementation of miR-148a-3p in progenitor cells blocked the inhibitory effect of PGC1β-OT1 on adipocyte formation. Moreover, overexpression of Kdm6b restored the osteoblast differentiation which was inhibited by silencing of endogenous PGC1β-OT. Our studies provide evidences that the novel LncRNA PGC1β-OT1 reciprocally regulates adipogenic and osteogenic differentiation through antagonizing miR-148a-3p and enhancing KDM6B effect.
Similar content being viewed by others
Introduction
It is known that adipocyte and osteoblast are both derived from marrow stromal cells (MSCs); thus a reciprocal and inverse relationship exists between adipogenesis and osteogenesis [1, 2]. For example, age-related bone loss and osteoporosis has been associated with reduced numbers of osteoblasts and increased numbers of adipocytes [3, 4]. Therefore, elucidating the mechanisms by which MSCs differentiate into marrow adipocytes and bone-forming osteoblasts is of great importance.
Adipogenesis is largely governed by peroxisome proliferator-activated receptor γ (PPARγ), an adipocyte-enriched nuclear receptor that cooperates with CCAAT/enhancer-binding proteins (C/EBPs), including C/EBPα, C/EBPβ, and C/EBPγ, to activate the transcription of genes such as adipocyte fatty acid-binding protein/adipocyte P2 (Fabp4/aP2) and fatty acid translocase (FAT/CD36) that give rise to adipocyte phenotype [5, 6]. In contrast, osteoblast development requires the activation of Wnt/β-catenin signaling and the expression of several master transcription factors including runt-related transcription factor (Runx2), osterix (Osx), and Distal-less homeobox 5 [7,8,9,10,11].
Long noncoding RNAs (LncRNAs) are transcripts more than 200 nucleotides in length, often polyadenylated but lacking protein-encoding capacity [12]. Previous studies have indicated that LncRNAs are involved in a wide variety of biological processes including but not limited to X chromatin inactivation [13], cancer metastasis [14], cell differentiation [15,16,17,18], and reprogramming of induced pluripotent stem cells [19, 20]. Importantly, studies have shown that LncRNAs are able to regulate gene expression through diverse mechanisms. For example, LncRNAs may function as RNA decoys, titrating transcription factors away from their DNA targets by directly binding to them [21, 22]. Furthermore, LncRNAs can also modulate posttranscriptional events during gene expression, such as splicing [23], mRNA translation [24], and mRNA degradation [25]. In addition, some LncRNAs act as a molecular sponge for microRNAs, and thereby indirectly enhance protein expression of the target genes of the microRNAs [26,27,28]. Although several LncRNAs have been shown to be differentially regulated during adipogenesis or osteogenisis [29, 30], the roles of most LncRNAs in adipogenic and osteogenic differentiation are unknown.
In the current study, we have identified and named a novel LncRNA PGC1β-OT1, which reciprocally regulates adipocyte and osteoblast differentiation from progenitor cells in vitro and in vivo. Moreover, PGC1β-OT1 contains a functional miR-148a-3p binding site, through which PGC1β-OT1 physically associates with miR-148a-3p and regulates lysine-specific demethylase 6b (KDM6B). Of interest, PGC1β-OT1 regulates adipocyte and osteoblast differentiation through antagonizing miR-148a-3p.
Results
PGC1β-OT1 is a LncRNA whose expression changed during adipogenesis and osteogenesis
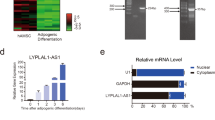
To explore whether LncRNAs are involved in the adipogenic differentiation of primary cultured MSCs, LncRNA profiles of the differentiated cells were analyzed 72 h after adipogenic treatment using Agilent mouse LncRNA 4× 180 K microarray. Differentially expressed LncRNAs were selected from microarray data for clustering analysis. It showed that 1113 annotated or potential LncRNAs were changed ≥3-fold in cells after adipogenic treatment vs. control cells (Fig. 1a, Supplementary Table S1). We focused on an uncharacterized RNA transcript PGC1β-OT1, which is located downstream of PGC1β and transcribed in the same direction as PGC1β (Fig. 1b). Analyses of the 5′- and 3′-rapid amplification of cDNA ends (RACE) showed that PGC1β-OT1 is 1759 nt in full length (Fig. 1c, S1). Bioinformatics analysis using coding potential calculator (CPC) indicated that PGC1β-OT1 lacks the ability to encode protein (Fig. 1d). qPCR analyses of fractionated nuclear and cytoplasmic RNA indicated that PGC1β-OT1 was localized in both the nuclear and cytoplasmic compartment. As control, 45S ribosomal RNA (rRNA) precursor was primarily localized in the nucleus, whereas 12S rRNA, a mitochondrial rRNA, was found in the cytoplasmic fraction (Fig. 1e). Furthermore, qRT-PCR showed that PGC1β-OT1 expression dramatically increased in stromal ST2 cells during adipogenesis, peaking at day 3 with 27-fold of increase (Fig. 1f). By contrast, it decreased at earlier stage (days 1–8) but increased at later stage (days 12–14) during osteogenic differentiation (Fig. 1g).
PGC1β-OT1 is a LncRNA whose expression changed during adipogenic and osteogenic differentiation. Hierarchical clustering of 1113 differentially (≥3-fold) expressed LncRNAs in primary marrow stromal cells 72 h after adipogenic treatment is shown (a). The location of PGC1β-OT1 on the Chromosome 18 is shown (b). 5′- and 3′-rapid amplification of cDNA ends and RT-PCR assays were performed to determine the full-length of PGC1β-OT1 (c). Coding potential calculator predicted the lack of coding ability of PGC1β-OT1. LncRNA H19 and protein-coding PGC1β and β-actin were also predicted as controls (d). qRT-PCR was performed to analyze PGC1β-OT1 levels in cytoplasmic (Cyt) and nuclear (Nuc) fractions (e). RNA expression profiles of PGC1β-OT1 during adipocyte (f) and osteoblast differentiation (g) are shown. Levels of PGC1β-OT1 in undifferentiated cells (day 0) are set as 1. Data are mean ± SD, n = 3. *p < 0.05 vs. Cyt (e) or vs. day 0 (f, g). LncRNA long noncoding RNA
PGC1β-OT1 negatively regulated adipocyte differentiation in progenitor cell lines
To study the functional role of PGC1β-OT1 during adipogenesis, we transfected the full-length PGC1β-OT1 expression construct into ST2. The overexpression of PGC1β-OT1 was verified by qRT-PCR (Fig. 2a). Notably, overexpression of PGC1β-OT1 in ST2 significantly decreased the number of differentiated oil red O-positive adipocytes (Fig. 2b, c). Accordingly, the mRNA levels of adipogenic transcription factors and marker genes, including C/EBPα, PPARγ, aP2 and adipsin were reduced in PGC1β-OT1 overexpressing ST2 cells vs. control, respectively, 48 h after adipogenic treatment (Fig. 2d). Consistently, the protein levels of C/EBPα, PPARγ and aP2 were also decreased in PGC1β-OT1 overexpressing ST2 cells 72 h after adipogenic treatment (Fig. 2e).
PGC1β-OT1 negatively regulated adipocyte differentiation in stromal ST2 cells. qRT-PCR analysis verified the overexpression of PGC1β-OT1 (a) or knockdown of PGC1β-OT1 (f) in ST2. Effects of PGC1β-OT1 overexpression (b−e) or silencing (g−j) on adipocyte differentiation were examined. Differentiated adipocytes were stained with oil red O (b, g). Oil red O extracted with isopropanol was measured at OD520 (c, h). The mRNA (d, i) and protein (e, j) levels of adipogenic factors were examined. Magnification, ×200. Data are mean ± SD, n = 3. *p < 0.05 vs. vector or small-interfering RNA negative control (NC)
Next, we used small-interfering RNAs (siRNAs) to silence PGC1β-OT1, followed by adipogenic treatment in ST2. Silencing of PGC1β-OT1 with two independent siRNAs markedly reduced the endogenous PGC1β-OT1 levels in ST2 (Fig. 2f), suggesting that the siRNAs work well in these cells. Transfection of the PGC1β-OT1 siRNAs promoted the differentiation of ST2 cells into adipocytes, as evidenced by the increase in the number of differentiated adipocytes (Fig. 2g, h), and enhanced expression of adipogenic transcripts of the adipogenic factors 48 h after adipogenic treatment (Fig. 2i). The protein levels of the adipogenic factors were also increased in the silencing cells 72 h after adipogenic treatment (Fig. 2j).
We also overexpressed PGC1β-OT1 in mesenchymal C3H10T1/2 cells (Figure S2A). Overexpression of PGC1β-OT1 inhibited formation of adipocytes (Figure S2B, C), and downregulated the mRNA and protein levels of adpogenic factors (Figure S2D, E). Conversely, we also silenced the endogenous expression of PGC1β-OT1 in C3H10T1/2 (Figure S2F). In presence of adipogenic medium, silencing of PGC1β-OT1 promoted adipocyte formation (Figure S2G, H), and upregulated the mRNA and protein levels of the adipogenic factors (Figure S2I-J).
Moreover, we tested whether silencing of PGC1β-OT1 spontaneously affects adipocyte differentiation. As shown in Figure S3, in absence of adipogenic medium, PGC1β-OT1 siRNAs stimulated adipogenic differentiation of C3H10T1/2 cells 14 days after transfection (Figure S3A, B). Consistently, PGC1β-OT1 siRNAs upregulated the mRNA and protein levels of the adipogenic factors (Figure S3C, D).
PGC1β-OT1 positively regulated osteoblast differentiation in progenitor cell lines
To further verify whether PGC1β-OT1 can regulate osteoblast differentiation, we transfected the full-length PGC1β-OT1 expression construct in ST2 cells followed by osteogenic treatment. Our results showed that overexpression of PGC1β-OT1 enhanced alkaline phosphatase (ALP) staining (Fig. 3a), and increased the mRNA levels of osteogenic factors, including Alp, Runx2, Osterix (Osx), and Osteopontin (Opn) 72 h after osteogenic treatment (Fig. 3b) in ST2 cells. Consistently, the protein levels of Runx2, Osterix, and ALP were increased in the PGC1β-OT1 overexpressing cells 72 h after osteogenic treatment (Fig. 3c).
PGC1β-OT1 positively regulated osteoblast differentiation in stromal ST2 cells. Effects of PGC1β-OT1 overexpression (a−c) or silencing (d−f) on osteoblast differentiation were examined in ST2. Differentiated osteoblasts were subjected to alkaline phosphatase staining (a, d). The mRNA (b, e) and protein (c, f) levels of osteogenic factors were examined. Data are mean ± SD, n = 3. *p < 0.05 vs. vector or NC
Subsequently, we tested whether silencing of endogenous PGC1β-OT1 alters osteoblast differentiation. As expected, knockdown of PGC1β-OT1 with two independent siRNAs inhibited osteoblast differentiation, as evidenced by the repressed ALP staining (Fig. 3d) and the downregulated mRNA levels of Alp, Opn, Bone sialoprotein (Bsp) and Osteocalcin (Bglap) (Fig. 3e) in ST2 cells. Moreover, the protein levels of Runx2, Osterix, and ALP were decreased in the silencing cells 72 h after osteogenic treatment (Fig. 3f).
We also tested if PGC1β-OT1 functions in preosteoblast MC3T3-E1 cells. The results showed that following osteogenic treatment, overexpression of PGC1β-OT1 promoted osteogenic differentiation, and increased the mRNA and protein levels of osteogenic factors in MC3T3-E1 cells (Figure S4A-D). Conversely, silencing of PGC1β-OT1 inhibited osteogenic differentiation and decreased the mRNA and protein levels of osteogenic factors in MC3T3-E1 cells (Figure S4E-H). Taken together, these data suggest that PGC1β-OT1 positively induces osteoblast differentiation.
PGC1β-OT1 regulated differentiation of primary MSCs in vitro and in vivo
To further determine the role of PGC1β-OT1 in differentiation of progenitor cells, we infected the primary cultured mouse MSCs with either control or PGC1β-OT1 shRNA lentivirus. Fluorecence observation revealed equal infection efficiency in the two groups of cells (Fig. 4a). qRT-PCR verified a significant decrease of PGC1β-OT1 expression in the cells infected with the shRNA virus (Fig. 4b). Following adipogenic treatment, silencing of endogenous PGC1β-OT1 promoted the formation of adipocytes from primary MSCs (Fig. 4c, d), and enhanced mRNA and protein expression of adipogenic factors (Fig. 4e, f). In contrast, silencing of PGC1β-OT1 inhibited osteoblast differentiation, as evidenced by attenuated ALP staining (Fig. 4g), and downregulated mRNA and protein levels of osteogenic factors as compared to control cells (Fig. 4h, i).
PGC1β-OT1 regulated differentiation of primary MSCs in vitro and in vivo. Fluorescence observation was performed to verify infection efficiency in primary MSCs (a). qRT-PCR was performed to verify PGC1β-OT1 knockdown after infection (b). Effects of PGC1β-OT1 silencing on adipogenic (c−f) and osteogenic differentiation (g−i) were examined. Differentiated adipocytes were stained with oil red O (c). Re-dissolved oil red O was measured at OD520 (d). The mRNA (e) and protein (f) levels of adipogenic factors were examined. Differentiated osteoblasts were subjected to alkaline phosphatase staining (g). The mRNA (h) and protein (i) levels of osteogenic factors were examined. qRT-PCR was done to verify the silencing of PGC1β-OT1 in MSCs of mice (j). Hematoxylin and eosin staining was done (k). Numbers (l) and areas (m) of adipocytes were quantified. Representative images of osteocalcin immunohistochemical staining are shown (n, Top: trabeculae; Bottom: endosteum). Numbers of osteoblasts on bone surface of trabeculae (o) and endosteum (p) in metaphysis were quantified. Data are mean ± SD. (a−i), n = 3. *p < 0.05 vs. Lenti-Ctrl. (j−p), n = 6. *p < 0.05 vs. NC. MSC marrow stromal cell
Furthermore, we investigated the physiological role of PGC1β-OT1 in mice, 2′-Ome-modified siRNA of PGC1β-OT1 was administrated into the bone marrow with an in vivo transfection reagent. qRT-PCR analysis confirmed that 3 days after intra-tibial injection, 2′-Ome-modified siRNA significantly downregulated the level of PGC1β-OT1 in MSCs (Fig. 4j). Thirty days after administration, mice treated with PGC1β-OT1 siRNA formed more adipocytes (Fig. 4k–m) and less osteocalcin-positive osteoblasts on the trabecular and endosteal bone surfaces as compared with NC-treated mice (Fig. 4n–p). These results provided evidences that PGC1β-OT1 plays a role in regulating adipogenic and osteogenic commitment of MSCs in vitro and in vivo.
PGC1β-OT1 contained a functional miR-148a-3p binding site
We further explored the mechanism by which PGC1β-OT1 regulates differentiation of progenitor cells. Previous studies have revealed that LncRNAs may act as sponges to bind specific miRNAs and affect their function [27, 31]. In our study, bioinformatics analysis revealed several putative binding sites for microRNAs including miR-378a-5p and miR-148a-3p within PGC1β-OT1 sequence (Fig. 5a). One site with starting nucleotide at position 5 nt is a 7-mer binding sequence complementary to the seed region of miR-378a-5p. The other site at position 172 nt has strong base-paring to the seed region of miR-148a-3p despite of a G:U pair.
PGC1β-OT1 contained functional miR-148a-3p binding site. Base pairing of PGC1β-OT1 with miR-378a-5p or miR-148a-3p is shown (a). Wild-type (WT-Luc) and mutant (Mutant-Luc) luciferase reporter constructs were made (b). miR-378a-5p mimics (c) or miR-148a-3p agomir (d) was cotransfected with WT-Luc or Mutant-Luc into HEK-293 cells and the luciferase activity was assayed. WT or mutant PGC1β-OT1 construct with mutation at miR-148a-3p binding site was transfected into ST2 cells, qRT-PCR was performed to verify the overexpression (e). Oil red O staining (f, g) and qRT-PCR analysis of adipogenic factors (h) were performed. Alkaline phosphatase staining (i) and qRT-PCR analysis of osteogenic factors (j) were performed. Data are mean ± SD, n = 3. *p < 0.05 vs. vector
To determine whether the miRNA binding sites are functional, we made a construct in which a mouse PGC1β-OT1 fragment was incorporated into pMIR-report vector at the 3′UTR of the luciferase gene (WT-Luc). This PGC1β-OT1 fragment was predicted to bind both miR-378a-5p and miR-148a-3p at position 5 and 172 nt, respectively. Alongside, we also made the mutant constructs in which the binding sequence of either miR-378a-5p or miR-148a-3p was mutated (Fig. 5b). As expected, miR-378a-5p mimics significantly reduced the luciferase activity of WT-Luc in HEK-293 cells compared to negative control mimics. To confirm that the observed inhibition was dependent on the predicted miR-378a-5p binding site, we tested the luciferase activity of the mutant construct. This mutant failed to respond to transfection of miR-378a-5p mimics (Fig. 5c). By contrast, the inhibition of miR-148a-3p agomir on the luciferase activity of WT-Luc was also observed, and the point mutation of WT-Luc at the binding sequence of miR-148a-3p rendered the construct to lose the response to miR-148a-3p agomir (Fig. 5d).
Subsequently, to further verify the necessity of the miRNA binding site for the function of PGC1β-OT1, we transfected into ST2 cells a WT or mutant PGC1β-OT1 with point mutation at the binding sequence of miR-378a-5p or miR-148a-3p. qRT-PCR verified the overexpression of the constructs (Fig. 5e). In presence of adipogenic medium, overexpression of the mutant PGC1β-OT1 for miR-148a-3p had no effect on adipocyte formation (Fig. 5f, g) and the expression levels of adipogenic factors (Fig. 5h) as compared to vector control. Furthermore, the mutant also failed to induce osteoblast differentiation (Fig. 5i, j). By contrast, overexpression of the mutant PGC1β-OT1 for miR-378a-5p blocked adipogenic differentiation and stimulated osteogenic differentiation as efficiently as the WT PGC1β-OT1 did (Figure S5). Together, these data suggest that the miR-148a-3p binding site within PGC1β-OT1 is required for the regulation of adipocyte and osteoblast differentiation.
PGC1β-OT1 physically associated with miR-148a-3p
To further determine whether PGC1β-OT1 may act as a sponge that physically associated with miR-148a-3p, in vivo crosslinking combined with affinity purification experiments were performed using S1 aptamer-tagged PGC1β-OT1 (Fig. 6a). The construct PGC1β-OT1 (untagged), S1-PGC1β-OT1 (tagged) or S1-PGC1β-OT1-Mut (tagged) was transfected into C3H10T1/2 cells, followed by crosslinking and affinity purification. RNA was extracted and subjected to reverse transcription and qRT-PCR. Effective affinity purification of tagged PGC1β-OT1 or PGC1β-OT1-Mut was demonstrated with significant 8–9-fold enrichment of the tagged vs. untagged PGC1β-OT1, while such an enrichment was not observed with β-actin mRNA (Fig. 6b). miR-148a-3p was also enriched by ~17 fold in tagged compared to the untagged PGC1β-OT1. However, the enrichment was abolished when transfected with tagged PGC1β-OT1-Mut. In contrast, no enrichment of U6 in tagged PGC1β-OT1 or PGC1β-OT1-Mut was seen (Fig. 6c). Furthermore, cotransfection of S1-PGC1β-OT1 with miR-148a-3p antagomir did not affect the enrichment of tagged PGC1β-OT1, but led to significantly reduced enrichment of miR-148a-3p as compared to the cotransfection of S1-PGC1β-OT1 with control antagomir (Fig. 6d, e). Together, these data suggest that PGC1β-OT1 physically associates with miR-148a-3p in vivo through the functional miR-148a-3p binding site.
PGC1β-OT1 physically associated with miR-148a-3p. Schematic outline of purification of PGC1β-OT1-associated microRNA ribonucleoprotein complex and RNA component identification (a). After transfection of single construct, in vivo crosslinking was done followed by affinity purification of PGC1β-OT1-associated miRNAs. Relative abundance of PGC1β-OT1, PGC1β-OT1-mut, and β-actin RNA (b) or relative abundance of miR-148a-3p and U6 (c) associated with tagged vs. untagged PGC1β-OT1 are plotted as relative fold enrichment. After cotransfection, in vivo crosslinking was done followed by affinity purification, and RNAs were extracted for qRT-PCR. Relative abundance of PGC1β-OT1 and β-actin RNA (d) or relative abundance of miR-148a-3p or U6 (e) associated with tagged vs. untagged PGC1β-OT1 are plotted as relative fold enrichment. Data are mean ± SD, n = 3. *p < 0.05 vs. untagged PGC1β-OT1, #p < 0.05 vs. tagged PGC1β-OT1
PGC1β-OT1 affected expression of endogenous miR-148a-3p and its target KDM6B
To determine whether PGC1β-OT1 regulation of miR-148a-3p is physiologically relevant, ST2 cells were induced with adipogenic treatment for 7 days. As shown in Fig. 7a, PGC1β-OT1 expression was strongly induced during adipogenic differentiation. A concomitant induction of miR-148a-3p was also observed. Intriguingly, PGC1β-OT1 overexpression significantly decreased the level of miR-148a-3p (Fig. 7b), suggesting that PGC1β-OT1 not only physically associates with miR-148a-3p, but also inhibits endogenous miR-148a-3p expression.
PGC1β-OT1 affected expression of endogenous miR-148a-3p and its target KDM6B. The expression profiles of PGC1β-OT1 and miR-148a-3p during adipogenesis were analyzed by qRT-PCR (a). Expression levels of PGC1β-OT1 and miR-148a-3p were examined after transfection of PGC1β-OT1 into ST2 (b). The mRNA (c) and protein (d, e) levels of KDM6B were determined after transfection of PGC1β-OT1, Mut-PGC1β-OT1 or vector into ST2 cells. The protein level of KDM6B was determined in ST2 after transfection of miR-148a-3p or control antagomir (f). Data are mean ± SD, n = 3. *p < 0.05 vs. vector or control antagomir
To further verify whether PGC1β-OT1 affects the expression of miR-148a-3p target, construct of PGC1β-OT1 or Mut-PGC1β-OT1 was transfected into ST2, and the expression of one known miR-148a-3p target KDM6B [32] was analyzed. The mRNA level of KDM6B was not changed in the PGC1β-OT1- or Mut-PGC1β-OT1-transfected cells (Fig. 7c). However, overexpression of PGC1β-OT1 increased the protein level of KDM6B, whereas the increase was abolished in the Mut-PGC1β-OT1-transfected cells (Fig. 7d, e). By contrast, the transfection with miR-148a-3p antagomir led to a significant increase of KDM6B protein (Fig. 7f). Together, these results strongly point to the notion that de-repression of endogenous miR-148a-3p on KDM6B was due to a direct inhibitory effect of PGC1β-OT1 on miR-148a-3p.
PGC1β-OT1 regulated differentiation of progenitor cells via its downstream effectors
To determine whether PGC1β-OT1 regulates adipogenic and/or osteoblast differentiation by affecting miR-148a-3p or its target gene KDM6B, on one hand, we undertook miR-148a-3p gain-of-function studies under the background of PGC1β-OT1 overexpression. The data revealed that the stimulatory effect of miR-148a-3p agomir on adipocyte formation was largely attenuated when PGC1β-OT1 was co-transfected into ST2 cells (Fig. 8a, b). Consistently, the mRNA levels of the adipogenic factors in the co-transfected cells were decreased compared to the cells transfected with miR-148a-3p agomir plus vector (Fig. 8c). On the other hand, we undertook KDM6B gain-of-function studies in ST2 under the background of PGC1β-OT1 silencing. The efficiency of Kdm6b overexpression and PGC1β-OT1 silencing was verified by qRT-PCR (Fig. 8d). The subsequent functional studies showed that the inhibitory effect of PGC1β-OT1 siRNA on osteoblast differentiation and expression of osteogenic factors was restored when Kdm6b expression construct was cotransfected (Fig. 8e, f).
PGC1β-OT1 regulated differentiation of progenitor cells via its downstream effectors. PGC1β-OT1 construct or the vector was cotransfected with miR-148a-3p or control agomir into ST2, and differentiated adipocytes were stained by oil red O (a). Oil red O extracted with isopropanol was measured at OD520 (b). The mRNA levels of adipogenic factors were examined (c). qRT-PCR analysis was performed to verify the silencing of PGC1β-OT1 and overexpression of Kdm6b (d). siPGC1β-OT1 or control small-interfering RNA was cotransfected with Kdm6b expression construct or vector, and differentiated osteoblasts were subjected to alkaline phosphatase staining (e). The mRNA levels of osteogenic factors were examined (f). Data are mean ± SD, n = 3. *p < 0.05 vs. NC plus Vector. #p < 0.05 vs. miR-148a-3p agomir plus Vector (b, c) or siPGC1β-OT1 plus Vector (f)
Discussion
There are several studies reporting expression and potential function of LncRNAs in adipocyte and/or osteoblast differentiation [29, 30, 33, 34]. However, a large portion of uncharacterized LncRNAs have not been identified. In the current study, a thousand of differentially expressed LncRNAs were found by microarray screening. Among these, we identified a novel LncRNA PGC1β-OT1, which is located downstream of PGC1β gene. We have shown that PGC1β-OT1 expression was dramatically altered during adipocyte and osteoblast differentiation. The data suggest that PGC1β-OT1 might play a role in the regulation of adipogenic and/or osteogenic differentiation from progenitor cells.
Based on the increase in the expression level of PGC1β-OT1 during adipogenesis, we reasoned that it might positively regulate adipogenesis. Unexpectedly, the data showed that PGC1β-OT1 overexpression in undifferentiated progenitor cells inhibited the formation of adipocytes, whereas silencing of PGC1β-OT1 induced formation of adipocytes. This pattern is similar to the case of LncRNA H19, which was strongly induced during myoblast differentiation, but showed inhibitory effect on muscle differentiation via antagonizing let-7 activity [31].
Given that a reciprocal and inverse relationship exists between adipogenesis and osteogenesis, we further examined the role of PGC1β-OT1 in osteoblast differentiation. The gain-of-function and loss-of-function studies demonstrated that PGC1β-OT1 positively regulated osteoblast differentiation. These findings provide evidence that PGC1β-OT1 may function to balance the reciprocal commitment of progenitor cells to adipocyte and osteoblast in vitro. Of more importance, in vivo experiments showed that silencing of endogenous PGC1β-OT1 significantly promoted the fat accumulation and attenuated the osteoblast formation in bone marrow of mice. The significant results have been observed at 1 month after only two injections although obvious change in trabecular bone volume was not observed at this time point (data not shown). These may define the physiological role of PGC1β-OT1 as a regulator of osteoblast and adipocyte differentiation, key processes in the maintenance and function of bone structure and the bone marrow microenvironment.
A number of LncRNA have been demonstrated to be primarily localized within the cytoplasm or shuttle between the nucleus and cytoplasm [35]. These RNAs function in diverse ways, interacting with a variety of protein-binding partners [36]. Furthermore, a growing body of evidences suggest that cytoplasmic LncRNAs can function as decoys for other noncoding RNAs such as miRNAs, and have thus been termed competing endogenous RNAs (ceRNA) [37, 38]. Recently, a new class of ceRNAs, called circular RNAs, has also been demonstrated to act as potent miRNA sponges [39]. In this study, we found that PGC1β-OT1 is localized both in the cytoplasm and nucleus, which suggests a possible mechanism that PGC1β-OT1 might be a ceRNA and function as a “sponge” to sequester miRNAs.
To further elucidate the molecular mechanisms by which PGC1β-OT1 regulates adipogenesis and osteogenesis, we searched for potential target miRNAs using a web-based program RNAhydrid. Two potential miRNAs that function in regulating adipocyte or osteoblast were predicted, i.e., miR-148a-3p and miR-378a-5p. miR-148a-3p has the ability to regulate adipocyte and osteoblast differentiation through targeting KDM6B [32], a recently identified key player in the development of both cartilage and bone [40]; miR-378a-5p, on the other hand, has been implicated in adipogenesis and lipogenesis [41]. Using luciferase reporter assays, we demonstrated that PGC1β-OT1 is capable of binding either miR-148a-3p or miR-378a-5p via the putative binding sequences that are complementary to the seed region of the respective microRNA. We then asked if the binding sequences on PGC1β-OT1 are functional in regulating directed differentiation of progenitor cells. We examined the function of the mutant PGC1β-OT1 constructs. The data showed that mutation of miR-148a-3p binding site rather than that of miR-378a-5p binding site rendered the PGC1β-OT1 construct to lose the capability in modulating adipogenic and osteogenic commitment of the progenitor cells. In addition, the crosslinking affinity purification study revealed an in vivo physical interaction between PGC1β-OT1 and miR-148a-3p. Supplementation of miR-148a-3p in progenitor cells blocked the inhibitory effect of PGC1β-OT1 on adipocyte formation. These data suggest that PGC1β-OT1 regulates adipogenic and/or osteogenic differentiation through binding and antagonizing the role of miR-148a-3p.
KDM6B, also known as Jumonji domain-containing 3 (Jmjd3), is a histone demethylase that specifically catalyzes the removal of trimethylation marks of histone H3K27 (H3K27me3). Ye et al. has for the first time identified KDM6B as a key player in osteogenic commitment of human MSCs by removing H3K27me3 at the promoter of Hox genes [40]. Our group has recently demonstrated that KDM6B acts as a reciprocal player in adipogenesis and osteogenesis in murine progenitor cells [42]. Moreover, we have demonstrated that miR-148a-3p plays a proadipogenic and antiosteogenic role through targeting and therefore negatively regulating the KDM6B demethylase [32].
In the present study, we further explored if there really exists a link between PGC1β-OT1 and KDM6B. The data showed that PGC1β-OT1 increased the protein level of KDM6B, but did not affect Kdm6b at the mRNA level. Moreover, overexpression of Kdm6b can attenuate the inhibitory effect of PGC1β-OT1 siRNA on osteoblast differentiation. Thus, bridged by miR-148a-3p, KDM6B is linked to PGC1β-OT1 and mediates the regulation of adipogenic and osteogenic commitment of MSCs by the latter. Given that KDM6B is crucial for macrophage differentiation, lung development and neurogenesis in vivo [42,43,44], we postulate that PGC1β-OT1/ miR-148a-3p axis may also contribute to these developmental processes.
In summary, we have identified a novel LncRNA PGC1β-OT1 that functions as a reciprocal modulator of adipocyte and osteoblast differentiation from progenitor cells, key processes in the maintenance and function of bone structure and bone marrow microenvironment. The function is based upon its binding to the proadipogenic and antiosteogenic microRNA miR-148a-3p. PGC1β-OT1 antagonizes the function of miR-148a-3p and thus potentiates the role of KDM6B in cell fate decision of marrow progenitor cells. Therefore, PGC1β-OT1 may play a fundamental role in bone homeostasis.
Materials and methods
Cell cultures
ST2, C3H10T1/2, and MC3T3-E1 cells were maintained in Dulbecco’s Modified Eagle Media (DMEM) containing 10% fetal bovine serum (FBS). For adipogenic differentiation, ST2 or C3H10T1/2 cells at 100% confluence were treated for 3 d with adipogenic medium (α-MEM containing 10% FBS, 0.5 μM dexamethasone, 0.25 mM methylisobutylxanthine, 5 μg/ml insulin, and 50 μM indomethacin), then for additional 2 d with 5 μg/ml insulin alone. The cells were allowed to fully differentiate for 2 more days in presence of adipogenic medium, followed by oil red O staining. For osteogenic differentiation, ST2 or MC3T3-E1 cells were cultured in osteogenic medium (α-MEM containing 10% FBS, 50 μg/ml ascorbic acid, and 5 mM β-glycerophosphate) for 3 d followed by RNA and protein isolation, or for 14 d followed by ALP staining.
Primary MSCs were collected from femurs and tibias of 4-week-old C57BL/6J mice and grown in α-MEM containing 10% FBS in a 10-cm dish. About 72 h after plating, supernatant containing nonadherent cells was removed and fresh medium was added. When grown to near confluence, the cells were passaged 2−3 times. The cells at passage 3–5 were treated with adipogenic or osteogenic medium to induce differentiation. All the experiments involving animals were carried out following the Chinese guidelines for animal welfare and experimental protocol, and was approved by the Animal Ethics Committee of Tianjin Medical University.
Quantitative RT-PCR
For mRNA study, RNA was extracted using a total RNA isolation kit (Omega Bio-Tek, Norcross, GA, USA). One microgram RNA was reverse transcribed into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Rockford, IL, USA). For miRNA study, RNA was extracted using an miRNA isolation kit (Omega Bio-Tek, Norcross, GA, USA), followed by reverse transcription using stem-loop primers. All the reverse transcription and PCR primers used are listed in Supplementary Table S2. Subsequently, cDNA was PCR-amplified with a SYBR Green fluorescence PCR kit (Sangon BioTech, Shanghai, China) on a real-time PCR cycler. The relative expression levels of the target genes were normalized against β-actin (for mRNA) or U6 (for miRNA) and determined as 2−ΔΔCT.
LncRNA array analysis
Primary MSCs were cultured in adipogenic medium for 3 days followed by total RNA extraction and purification. The total RNA was amplified and labeled by using Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies, Santa Clara, CA, USA). Then the labeled cRNA was purified by RNeasy mini Kit (QIAGEN, GmBH, Germany). The LncRNA expression profiles were analyzed using Agilent Mouse LncRNA 4× 180 K Microarray (Design ID: 046161). Data were extracted with Feature Extraction software 10.7 (Agilent Technologies, Santa Clara, CA, USA). Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent Technologies, Santa Clara, CA, USA). Differential expression LncRNAs (fold change ≥ 3) were selected from microarray data for clustering analysis.
Rapid amplification of cDNA ends (RACE)
5′- and 3′-RACE were performed using the SMARTer RACE 5′/3′ Kit (Clontech, Palo Alto, CA, USA). Briefly, 1 μg total RNA from each sample was converted into 5′- or 3′-RACE-Ready first-strand cDNA using a 5′CDS primer A or a 3′ CDS primer (included in the kit), respectively. The cDNA was amplified using a universal primer (UPM, 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′) mixed with 5′ or 3′ gene-specific primer: 5′GSP: 5′-GATTACGCCAAGCTTAATCCCAGGCACAGTAATCAG-3′; 3′GSP: 5′-GATTACGCCAAGCTTAACGTACTGCTTCCTTGATTG-3′. Thereafter, the PCR products were used as the template for the nested PCR with the following primers: Universal Primer Short, 5′-CTAATACGACTCACTATAGGGC-3′; 5′NGSP, 5′-GATTACGCCAAGCTTACTATATCCTCTTTCCCCGTGG-3′; 3′NGSP, 5′-GATTACGCCAAGCTTGCCCCTCCCACCCGACTGACTTCCTTCT-3′. The amplified cDNA were cloned into the pRACE vector and sequenced.
Constructs
The full-length PGC1β-OT1 cDNA was PCR-amplified with ST2 cDNA as a template using the following primers: forward, 5′-GTCGGATCCTGATGACATT GTTACTTGTG-3′, and reverse, 5′-GGAATTCGATCAGGTTATGACAGGAACGG CC-3′. The PCR product was restricted with BamHI/EcoRI sites and then cloned into pcDNA3.1(+) vector.
To make mutant PGC1β-OT1 construct, PCR was performed using WT-PGC1β-OT1 construct as a template by using a mutagenesis kit (Vazyme Biotech, Nanjing, China) to mutate the binding site for miR-378a-5p or miR-148a-3p respectively. To add S1 tag to the WT or mutant PGC1β-OT1, an annealed oligonucleotide fragment containing a 44-nt S1 sequence (sense: 5′-AGCTTACCGACCAGAATCATGCAAGTGCGTAAGATAGTCGCGGGCCGG-3′; antisense: 5′-GATCCCGGCCCGCGACTATCTTACGCACTTGCATGATTCTGGTCGGTA-3′) was inserted into the WT or mutant PGC1β-OT1 construct at HindIII/BamHI sites.
The luciferase construct of PGC1β-OT1 (WT-Luc) was created by inserting into pMIR-REPORT a 495-bp PCR fragment containing predicted binding sites of miR-378a-5p and miR-148a-3p using the following primers: forward, 5′-GACTAGTGTCGTCCTCGCTTCCAGC-3′; reverse, 5′-CAAGCTTTACAGCT CGAGTGGAACTCAC-3′. To make mutant reporter construct (Mutant-Luc), PCR-amplification was done using mutant PGC1β-OT1 as a template and the above-mentioned primers and then inserted into pMIR-REPORT.
Transfections
For the PGC1β-OT1 gain-of-function experiments, ST2, C3H10T1/2 or MC3T3-E1 cells were transfected for 18 h with PGC1β-OT1 expression plasmid or the vector using X-tremeGene HP transfection reagent (Roche Applied Science, Mannheim, Germany). For PGC1β-OT loss-of-function studies, the cells were transfected for 18 h with either 30 nM PGC1β-OT1 siRNAs or control siRNA (Genepharma, Shanghai, China) using lipofectamine RNAimax (Invitrogen, Carlsbad, CA, USA). The sequences of the siRNAs are listed in Supplementary Table S3. For the cotransfection experiments, lipofectamine 3000 was used to transfect PGC1β-OT1 expression plasmid along with miR-148a-3p agomir, or PGC1β-OT1 siRNA along with KDM6B construct for 18 h. Adipogenic or osteogenic induction was performed at appropriate confluence to allow the cells to differentiate.
Luciferase reporter assay
The assays were carried out in a 24-well plate as previously described [45]. Briefly, 500 ng of WT-Luc or Mut-Luc construct was transfected into HEK-293 cells, together with control miRNA, miR-148a-3p agomir or miR-378a-5p mimics (Genepharma, Shanghai, China) at a final concentration of 30 nM. Luciferase activity was measured 48 h posttransfection using Dual-Luciferase Reporter Assay System (E1960, Promega, Madison, WI, USA). Firefly luciferase activity was normalized against Renilla luciferase activities.
In vivo formaldehyde crosslinking and affinity purification
C3H10T1/2 cells were transfected with PGC1β-OT1, S1-PGC1β-OT1, or S1-PGC1β-OT1-mut in a 10-cm plate. Cells were harvested 48 h after transfection. Crosslinking was done in Phosphate Buffered Saline (PBS) containing 0.37% formaldehyde and then stopped by addition of 0.25 mol/L glycine. Cell pellet was lysed and then incubated with 200 μl of Streptavidin MagneSphere (Z5481, Promega, Madison, WI, USA). The beads were collected, washed, eluted with elution buffer and then heated at 70 °C to reverse crosslinks. Then RNA was extracted using TRIzol reagent (Invitrogen).
Lentiviral packaging and infection
To make the lentiviral construct coding for PGC1β-OT1 shRNA, we synthesized a pair of oligos against PGC1β-OT1 with the following sequences: forward, 5′-GATCCGCCATATCCAGTGTGACATTTCAAGAGAATGTTCACACTGGATATGGCTTTTTTG-3′, reverse, 5′-AATTCAAAAAAGCCATATCCAGTGTGACATT CTCTTGAAATGTCACACTGGATATGGCG-3′. The oligo pair was annealed and then inserted into the pLVX-shRNA2 vector (Clontech, Mountain View, CA, USA). The shRNA or control lentiviruses were packaged by transfecting 293T cells with the lentiviral shRNA plasmid or the empty vector using a packaging system (Jiman Biotech, Shanghai, China) following the supplier’s protocol. The supernatant containing the viruses was harvested, concentrated and titered. The viruses were used at an MOI (multiplicity of infection) of 20 to infect primary MSCs. The adipogenic or osteogenic medium was added at appropriate confluence to allow the cells to differentiate.
In vivo delivery of siRNA
A total of 30 male, 6-week-old C57BL/6 mice were purchased from SFP (Beijing) Biotechnology (Beijing, China). The mice were housed with controlled room temperature (22 ± 2 °C) and humidity (55 ± 5%) with a 12/12 h light−dark cycle and free access to food and water.
2′-Ome-modified PGC1β-OT1 siRNA (siRNA2) and control siRNA were purchased from Genepharma (Shanghai, China). 10 micrograms siRNA was diluted in 12.5 μl 5% glucose solution. In a separate tube, 1.2 μl in vivo-jetPEI (N/P ratio = 6, Catalogue no. 201-10 G, Polyplus-transfection S.A, illkirch, France) was diluted in 12.5 μl 5% glucose solution, then mixed with the siRNA solution. The mixed solution (25 μl) was incubated for 15 min at room temperature.
Mice received intra-tibial injection of above-mentioned 25 μl PGC1β-OT1 siRNA or control siRNA solution, twice with an interval of 2 weeks. The other side tibia was injected with an equal volume of PBS alone. Tissues were collected after sacrifice at day 3 and day 30 post-injection. For day 3 samples, MSCs were collected from tibias and femurs, and then grown on a 10-cm dish for 48 h, followed by RNA isolation and qRT-PCR. For day 30 samples, tibias were harvested, fixed in 10% formalin for 3 days, and decalcified in 14% Ethylenediaminetetraacetic acid (EDTA) for 14 days, followed by embedding in paraffin. Five-micrometer-thick longitudinally oriented bone sections were stained with hematoxylin and eosin (H/E). The region of interest selected for analysis is 1 mm in length from 100 μm below the growth plate in proximal tibial metaphysis to determine the number and area of adipocytes and number of osteocalcin-positive osteoblasts.
Immunohistochemical staining
Immunohistochemical staining was performed as previously described with minor modification [46]. In brief, bone sections were digested with 0.05% trypsin at 37 °C for 20 min for antigen retrieval, and then incubated with primary antibody against osteocalcin (Proteintech, Wuhan, China) at 4 °C overnight. Subsequently, an HRP-DAB system (Proteintech, Wuhan, China) was used to detect the immunoactivity, followed by counterstaining with hematoxylin.
Bioinformatics analysis
miRNA binding sites on PGC1β-OT1 were predicted using a web-based program RNAhydrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/). Transcription potential measurement was performed using CPC (http://cpc.cbi.pku.edu.cn/).
Western blot analysis
Cells were lysed, and proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were incubated overnight with primary antibodies including rabbit mAbs by Abcam (Cambridge, MA, USA): anti-C/EBPa, and anti-ALP; rabbit mAb by Cell Signaling Technology (Danvers, MA, USA): anti-PPARγ; mouse mAb by MBL (Nagoya, Japan): anti-Runx2; rabbit polyclonal antibody by Abcam: anti-osterix and anti-KDM6B; rabbit polyclonal antibodies by Proteintech (Wuhan, China): anti-aP2 and anti-β-actin. Then the membranes were incubated with the corresponding horseradish peroxide-labeled IgG (1:3000) for 2 h. Finally chemiluminescence reagent (Advansta, Menlo Park, CA, USA) was used to visualize the results.
Oil red O staining
Fully differentiated adipocytes were fixed in 4% paraformaldehyde for 10 min. The samples were washed with water and then stained in 60% saturated oil red O solution for 5 min. For oil red O quantification, isopropanol was added to dissolve the stain and light absorbance was measured at 520 nm.
ALP staining
Cells cultured for 14 d were fixed in 10% formalin for 10 min and stained with the 1-Step nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) staining kit (Pierce, Thermo Scientific, Rockford, IL, USA) for 15 min.
Statistical analysis
Data are expressed as the mean ± SD. For the relative mRNA, miRNA, protein and luciferase activity quantification, the means of the control groups are set to 1. For two-group comparison, Student’s t test was performed. For multiple-group comparison, one-way ANOWA analysis was performed. If the one-way ANOVA was significant, a post-hoc comparison was performed using the Newman−Keuls test. A p value of <0.05 was considered to be statistically significant.
References
Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–66.
Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–39.
Idris AI, Sophocleous A, Landao-Bassonga E, Canals M, Milligan G, Baker D, et al. Cannabinoid receptor type 1 protects against age-related osteoporosis by regulating osteoblast and adipocyte differentiation in marrow stromal cells. Cell Metab. 2009;10:139–47.
Yu B, Wang CY. Osteoporosis: the result of an ‘aged’ bone microenvironment. Trends Mol Med. 2016;22:641–4.
Kawai M, Rosen CJ. PPARgamma: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol. 2010;6:629–36.
Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–8.
Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–38.
Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8.
Artigas N, Gamez B, Cubillos-Rojas M, Sanchez-de Diego C, Valer JA, Pons G, et al. p53 inhibits SP7/Osterix activity in the transcriptional program of osteoblast differentiation. Cell Death Differ. 2017;24:2022–31.
Hojo H, Ohba S, He X, Lai LP, McMahon AP. Sp7/Osterix is restricted to bone-forming vertebrates where it acts as a Dlx co-factor in osteoblast specification. Dev Cell. 2016;37:238–53.
Rattanasopha S, Tongkobpetch S, Srichomthong C, Kitidumrongsook P, Suphapeetiporn K, Shotelersuk V. Absent expression of the osteoblast-specific maternally imprinted genes, DLX5 and DLX6, causes split hand/split foot malformation type I. J Med Genet. 2014;51:817–23.
Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176.
Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166–015-0733-y.
Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28:287–301.
Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen H, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun. 2017;8:14718.
Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–47.
Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16:318–25.
Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–83.
Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev Cell. 2017;42:333–348.e5.
Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev. 2013;22:2240–53.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP, Noncoding RNA. gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8.
Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–33.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38.
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55.
Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–8.
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H. et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118-017-0685-9
Li Z, Wu X, Gu L, Shen Q, Luo W, Deng C, et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis. 2017;8:e2888.
Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBasev2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–8.
Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Rep. 2015;5:856–65.
Chen J, Liu Y, Lu S, Yin L, Zong C, Cui S. et al. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int J Obes (Lond). 2017;41:299–308.
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12.
Tian L, Zheng F, Li Z, Wang H, Yuan H, Zhang X, et al. miR-148a-3p regulates adipocyte and osteoblast differentiation by targeting lysine-specific demethylase 6b. Gene. 2017;627:32–39.
Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-beta1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33:3481–92.
Jin C, Jia L, Huang Y, Zheng Y, Du N, Liu Y, et al. Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived stem cells. Stem Cells. 2016;34:2707–20.
Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genom Proteom Bioinforma. 2016;14:73–80.
Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–7.
Li MJ, Zhang J, Liang Q, Xuan C, Wu J, Jiang P, et al. Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res. 2017;45:5653–65.
Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell. 2014;56:347–59.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11:50–61.
Ishida M, Shimabukuro M, Yagi S, Nishimoto S, Kozuka C, Fukuda D, et al. MicroRNA-378 regulates adiponectin expression in adipose tissue: a new plausible mechanism. PLoS ONE. 2014;9:e111537.
Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, et al. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014;8:1290–9.
Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K, et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet. 2014;10:e1004524.
Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–44.
Zhou J, Guo F, Wang G, Wang J, Zheng F, Guan X, et al. miR-20a regulates adipocyte differentiation by targeting lysine-specific demethylase 6b and transforming growth factor-beta signaling. Int J Obes (Lond). 2015;39:1282–91.
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125:1509–22.
Acknowledgements
The work was supported by grants nos. 81601864, 81501846, 81672116, and 81472040 from National Natural Science Foundation of China. The work was also partially supported by grant no. 2015KYZM09 from the Scientific Foundation of Tianjin Medical University and by grant no. 2015RC02 from Key Lab of Hormones and Development (Ministry of Health).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by R.A. Knight
Supplementary information
Rights and permissions
About this article
Cite this article
Yuan, H., Xu, X., Feng, X. et al. A novel long noncoding RNA PGC1β-OT1 regulates adipocyte and osteoblast differentiation through antagonizing miR-148a-3p. Cell Death Differ 26, 2029–2045 (2019). https://doi.org/10.1038/s41418-019-0296-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-019-0296-7
This article is cited by
-
Oncostatin M receptor regulates osteoblast differentiation via extracellular signal-regulated kinase/autophagy signaling
Stem Cell Research & Therapy (2022)
-
LncRNA XIST accelerates burn wound healing by promoting M2 macrophage polarization through targeting IL-33 via miR-19b
Cell Death Discovery (2022)
-
LncRNA GACAT2 binds with protein PKM1/2 to regulate cell mitochondrial function and cementogenesis in an inflammatory environment
Bone Research (2022)
-
MiR-148a deletion protects from bone loss in physiological and estrogen-deficient mice by targeting NRP1
Cell Death Discovery (2022)
-
Long noncoding RNA LYPLAL1-AS1 regulates adipogenic differentiation of human mesenchymal stem cells by targeting desmoplakin and inhibiting the Wnt/β-catenin pathway
Cell Death Discovery (2021)