Abstract
Current literature agrees on the notion that efficient DNA repair favors longevity across evolution. The DNA damage response machinery activates inflammation and type I interferon signaling. Both pathways play an acknowledged role in the pathogenesis of a variety of age-related diseases and are expected to be detrimental for human longevity. Here, we report on the anti-inflammatory molecular make-up of centenarian’s fibroblasts (low levels of IL-6, type 1 interferon beta, and pro-inflammatory microRNAs), which is coupled with low level of DNA damage (measured by comet assay and histone-2AX activation) and preserved telomere length. In the same cells, high levels of the RNAseH2C enzyme subunit and low amounts of RNAseH2 substrates, i.e. cytoplasmic RNA:DNA hybrids are present. Moreover, RNAseH2C locus is hypo-methylated and RNAseH2C knock-down up-regulates IL-6 and type 1 interferon beta in centenarian’s fibroblasts. Interestingly, RNAseH2C locus is hyper-methylated in vitro senescent cells and in tissues from atherosclerotic plaques and breast tumors. Finally, extracellular vesicles from centenarian’s cells up-regulate RNAseH2C expression and dampen the pro-inflammatory phenotype of fibroblasts, myeloid, and cancer cells. These data suggest that centenarians are endowed with restrained DNA damage-induced inflammatory response, that may facilitate their escape from the deleterious effects of age-related chronic inflammation.
Similar content being viewed by others
Introduction
Human longevity represents the capability of exceptional individuals to reach the extreme limits of human lifespan while avoiding age-related diseases, such as atherosclerosis and cancer [1]. A common feature of age-related diseases is the activation of inflammatory pathways that recalls the systemic pro-inflammatory status that develops with age, named inflamm-aging [2]. Studies on centenarians convey that such long-living subjects escape the majority of age-related diseases and avoid the detrimental effects of inflamm-aging [1, 2]. An explanation of such phenomenon stands in the genetic and epigenetic make-up of such exceptional individuals that may restrain the activation of the innate immunity/inflammation in response to lifelong stress [2]. As it occurs across evolution, individuals that attain longevity are likely to be endued with a peculiar capability to repair DNA damage, thus maintaining genomic integrity lifelong [1, 2]. In our previous investigation, we showed that centenarians are endowed with well-preserved telomeres [3] and a highly efficient DNA repair response (DDR) mainly due to the activation of 53BP1-dependent non-homologous end joining (NHEJ) mechanism [4]. DNA damage is a powerful trigger of inflammation and type I interferon (IFNalpha/beta) signaling [5,6,7]. Recent literature shed light on the importance of cytoplasmic nucleic acid sensors in DNA-damage response [6]. Indeed, upon DNA damage, nucleic acids are misplaced into the cytoplasm, whereby they engage evolutionary-conserved sensors that trigger inflammation and IFNalpha/beta pathways [6, 7]. This mechanism is pathogenic in rare diseases called interferonopathies [8], but it is expected to be also linked to common age-related autoimmune diseases, such as systemic lupus erythematosus and psoriasis [8, 9]. The cytoplasmic misplacement of nucleic acids recently gained importance in explaining the onset of the senescent associated pro-inflammatory secretome [10]. In particular, it was demonstrated that the absence of the cytosolic DNA sensor cGAS incapacitates chronic DDR activation to elicit senescence [11]. The relationship between cell senescence and systemic aging is a major matter of debate. Indeed, senescent cells are likely to constitute a minor population in aged tissues [12]. However, owing to their capability of secreting a huge amount of inflammatory mediators they may contribute to systemic inflammation and thus to inflamm-aging [13]. Moreover, current literature pinpoints that the ablation/injection of few senescent cells is enough to reduce/increase progeric features at systemic level in the mice [14, 15]. Here, we hypothesize that centenarians may be endowed with a molecular setting that allows them to maintain genomic stability while avoiding the activation of DNA damage associated inflammatory/INFbeta response. To this purpose, we measured the level of DNA damage in dermal fibroblasts (DF) from centenarian’s cells under basal and upon stress (gamma radiations) and their capability to express interleukin-6 (IL-6, “the cytokine for gerontologist”) [16], a major predictor of morbidity and mortality in aged people [17]. In the same experimental setting, we measured INFbeta, owing to that type I interferon is triggered by DNA damage and it elicits systemic progeric features in the mouse model [18]. In particular, we then assessed the level of highly pro-inflammatory species such as cytoplasmic RNA:DNA hybrids [6, 19, 20] in fibroblasts of people of different ages and the expression of the cognate degrading enzyme RNAseH2 [8]. Indeed, the degradation of RNA:DNA hybrids is a crucial physiologic phenomenon to preserve normal cell functioning and RNA:DNA hybrids accumulation elicit INFalpha/beta signaling, genomic instability, and telomere derangement in a variety of cellular models [8, 21, 22]. Finally, owing to the fact that extracellular vesicles (EV) take part in inflammation and INFalpha/beta activation [23,24,25], we assessed the capability of EV from centenarian’s fibroblasts to modulate inflammatory activation in a variety of recipient cells.
Materials and methods
Cell cultures
Primary DF (Supplementary Table 1) were obtained from healthy young (18–40 years), old (58–98 years), and centenarian (>99 years, Supplementary Table 2) subjects. DF were cultured in 10% FCS DMEM (Euroclone, Milan, Italy). All the experiments were performed between the 3rd and 6th in vitro passage. DF were collected upon signed informed consent. The study was approved by the local Ethics Committee (S. Orsola-Malpighi Hospital Prot. n. 2006061707, amendment 08/11/2011; Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Prot. n. 2035, amendment 30/11/2011; University of Calabria 9/9/2004 amendment on 24/11/2011). Early passages and senescent human embryonic diploid lung fibroblasts were cultured as previously described [26]. THP1 cells were maintained in 20% FCS RPMI 1640 medium. Cells were seeded in 24-well plates at a density of 500.000 cells/ml. For M1–M2 monocyte polarization assay, THP1 were exposed to 1 × 103 CD9+ EV/cell for 72 h. MCF-7 cells were cultured in 10% RPMI 1640 medium. MCF-7 MS were generated by seeding single cell suspension (1–5 × 103 cells per well) in low attachment plates filled with MammoCult™ (Stem Cell Technologies, Vancouver, Canada) and assessed at day 7 [27]. HUVECs were cultured as previously described [28].
Exposure of DF to DNA damaging agents
DF were exposed to a single radiation dose of 7.5 Gy using the linear acceleration Elekta Synergy Platform system (Elekta Oncology Systems, Stockholm, Sweden) as previously described [29]. DNA damage was evaluated 72 h after treatment. Doxorubicin (Sigma, St. Louis, MO) was used at a concentration of 5 μM for 24 h.
Comet assay
The assay was performed according to the manufacturer’s protocol (Comet Assay, Trevigen, Gaithersburg, MD). Briefly, at the end of the treatments, 5 × 105 cells were suspended in LMA agarose (at 37 °C) at a ratio of 1:10 (v/v), and 75 μl were immediately transferred onto the comet slide. The slides were immersed for 1 h at 4 °C in a lysis solution, washed in the dark for 1 h at room temperature in alkaline solution, then electrophoresed for 30 min at 20 V. Slides were immersed twice in dH2O for 5 min each, then dipped in 70% ethanol and stained with the Silver Staining Kit (Trevigen). The extent of DNA damage was evaluated quantitatively by Olympus IX51 microscope 10×, Olympus Corporation, Tokyo, Japan, by scoring at least 100 nucleoids of different categories, using the free software Cell Profiler (version 2.1.1). Percentage of DNA in the tail for different categories of comets was expressed.
Confocal microscopy analysis
Cells were fixed and permeabilized with ice-cold methanol for 10′ and acetone for 1′ on ice, blocked with 2% BSA, and incubated with primary anti-S9.6 antibody (1:100 dilution, Kerafast, Boston, MA, USA) and secondary goat anti-mouse Alexa Fluor 488 (1:250; Life Technologies). Alternatively, cells were fixed with paraformaldehyde 4% for 20 min at room temperature and incubated overnight at 4 °C with anti Rad51 diluted 1:100 (Millipore, Milan, Italy). The confocal imaging was performed with a Nikon A1 confocal laser scanning microscope, equipped with a 60×, 1.4 NA objective and with 405, 488, and 561 nm laser lines. Rad51 foci were enumerated using Fiji (ImageJ).
Transient gene knock-down and over-expression
RNAseH2C (HSS149830) and Snai2/Slug (HSS109993/109995/185949) specific siRNA, the GC matched content control (siSCR) were transiently transfected with lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) following manufacturer instructions. All transfections were performed onto 70% confluent cells. The cells were harvested after 72 h of siRNA transfection and stored in TRIzol® (Life Technologies, Carlsbad, CA, USA).
Real-time PCR analysis
Total RNA was extracted from cells using TRIzol® (Life Technologies, Carlsbad, CA, USA). TaqMan probes for Rad51, Snai2/Slug, IL-6, RNAseH2A/B/C, interferon beta-1, TREX1, IL-1, PPRalpha, PPRgamma, RXRalpha, Alox-15, Jagged-1, IL-10, CD163, CD68, and glucuronidase were purchased from Life Technologies (Carlsbad, CA, USA). Quantitative PCR analysis was performed on Light-Cycler using iQ software (BIORAD, Hercules, CA, USA), mRNA levels were calculated with the 2−ddct method.
Telomere length assessment
Telomere length was measured as telomere-to-single copy gene ratio (T/S), using quantitative real-time PCR as previously described [30] with some modifications. For cellular samples, 5 µl aliquots containing 20 ng DNA and 10 µl of master mix per sample were processed. For EV samples, 5 µl aliquots containing 0.8 ng DNA and 10 µl of master mix per sample were processed. For each standard curve, one reference DNA sample was diluted serially in water by 1.68-fold dilution to produce 5 DNA concentrations ranging from 30 to 2 ng in 5 µl. To reduce inter-assay variability, the telomere and the single-copy gene (36B4) were analyzed on the same plate. Primer sequences and concentrations for telomere and 36B4 were as previously described [30]. The thermal cycling profile was: (1) one cycle of 10 s at 95 °C; (2) 30 cycles of 5 s at 95 °C, 15 s at 57 °C, and 20 s at 72 °C. Measurements were performed in duplicate and reported as T/S ratio relative to a calibrator sample (Roche, Milano, Italy) to allow comparison across runs. EV telomeric sequences were quantified based on the RT-PCR CT, without the comparison with single gene copy. To date no single gene copy was described in EV. The real-time Chromo4 MJ Research system (Bio-Rad Laboratories, Hercules, CA, USA) was used for all PCRs. The coefficients of variation within duplicates of the telomere and single-gene assay were 2% and 1.8%, respectively. Approximately 30% of the samples were repeated on different plates to assess T/S measurement reproducibility. The inter-assay coefficient of variation was <10%. All analyses were blind. The correlation coefficient between T/S and the telomere restriction fragment was R2 = 0.88
ELISA test
IL-6 ELISA was performed with Quantikine ELISA kit for human IL-6 (R&D System, Minneapolis, MN, USA) according to manufacturer instructions. Concentrations of Maresin 1 and Resolvin D1 (RvD1) in cell culture supernatants were measured using commercially available ELISA kits (Cayman Chemical Co., Ann Arbor, MI, USA) according to manufacturer instructions.
Western blot analysis
Proteins were extracted with radio-immunoprecipitation assay buffer, with the exception of γ-H2AX extraction which was performed by adding HCl to the lysis buffer [10 mmol/L HEPES (pH 7.9); 1.5 mmol/L MgCl2; 10 mmol/L KCl] to a final concentration of 0.2 mol/L. The following antibodies were used: anti-actin (C4—Santa Cruz, CA); anti-phospho-(gamma)-Histone H2AX (Ser139, clone JBW301, Millipore, Billerica, MA, USA); anti-Rad51, clone 3C10 (Millipore); anti-Snai2/Slug (L40C6, Cell Signalling, Beverly, MA, USA); anti-RNASeH2C (Protein-tech, IL, USA), and anti-actin (clone C4, Santa Cruz, CA, USA) as loading control.
DNA methylation analysis
DNA methylation of RNAseH2C locus (genomic region: chr11:65,486,750–65,487,179, GRCh37/hg19) was evaluated using the EpiTYPER assay (Agena Bioscience). Briefly, the EZ-96 DNA Methylation Kit (Zymo Research Corporation) was used to perform bisulfite-conversion of 500 ng of genomic DNA. This treatment specifically converts unmethylated cytosines to uracils, while methylated cytosines remain unaffected. Ten nanograms of converted DNA were amplified using the following primers for bisulfite-converted DNA: RNAseH2C_F_aggaagagagATGGGGTTGAGGATAGTTTAAAAAG; RNASEH2C_R_cagtaatacgactcactatagggagaaggctCCCAATAAAAAAACTCTTCACAACA. PCR products were then processed and analyzed according to EpiTYPER protocol. The following Gene Expression Omnibus datasets were analyzed for RNAseH2C methylation according to Illumina HumanMethylation450 BeadChip array measurements: GSE40279, which includes whole-blood DNA methylation data from 656 healthy subjects with age ranging from 19 to 101 years; GSE46394, which includes DNA methylation data from 15 atherosclerotic lesion tissues and matched normal aortic tissue, plus 19 carotid atherosclerotic samples; GSE66695, which includes DNA methylation data from 40 normal and 80 breast cancer samples.
Small RNA seq analysis
Small RNA libraries were prepared from 6 DF samples using TruSeq Small RNA Library PrepKit v2 (Illumina, RS-200-0012/24/36/48), according to manufacturer’s indications. Briefly, 35 ng of purified RNA were linked to RNA 3′ and 5′ adapters, converted to cDNA and amplified using Illumina primers containing unique indexes for each sample. Each library was quantified using Agilent Bioanalyzer and High Sensitivity DNA kit (Agilent, 5067-4626) and an equal amount of libraries were pooled together. A size-selection was performed to keep fragments between 130 and 160 bp. After ethanol precipitation, the library pool was quantified with Agilent High Sensitivity DNA kit, diluted to 1.8 pM and sequenced using NextSeq® 500/550 High Output Kit v2 (75 cycles) (Illumina, FC-404-2005) on Illumina NextSeq500 platform. Raw base-call data generated from the Illumina NextSeq500 system were demultiplexed by Illumina BaseSpace Sequence Hub (https://basespace.illumina.com/home/index) and converted to FASTQ format. After a quality check, which was performed with FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the adapter sequences were trimmed using Cutadapt (http://cutadapt.readthedocs.io/en/stable/index.html). In this step, sequences shorter than 10 nucleotides were also removed. Read mapping was performed using STAR algorithm (https://www.ncbi.nlm.nih.gov/pubmed/23104886). The reference genome was constituted by human microRNAs sequences from the miRbase 21 database (http://www.mirbase.org/). Counts of raw mapped reads were obtained using the htseq-count script from the HTSeq tools (http://www-huber.embl.de/HTSeq/doc/overview.html); raw counts were further normalized using DESeq2 bioconductor package (http://bioconductor.org/packages/release/bioc/html/DESeq2.html). Data were analyzed using Genespring GX software v. 14.8 (Agilent Technologies). Differentially expressed miRNAs were identified using moderated t-test (FDR 10% with Benjamini–Hochberg correction), principal component analysis and cluster analysis, with Manhattan correlation, were performed using GeneSpring GX software.
CD9+ exosome isolation and phenotypization from DF supernatants
CD9+ exosomes from young, old, and centenarians DF supernatants were isolated by CD9 immunobeads isolation kit (HansaBioMed Life Sciences Ltd.; cod: HBM-BOLC-CC/20-1) according to manufacturer’s protocol. EVs were diluted to approximately 1 ml of PBS, loaded into the sample chamber of an LM10 unit (Nanosight, Malvern, UK) and three videos of either 30 or 60 s were recorded of each sample. Exosomal-surface antigens were investigated with the MACSPlex Exosome kit (Miltenyi Biotec GmbH, Gladbach, Germany). Briefly, after isolation, exosomes were diluted in MACSPlex buffer and stained according to manufacturer instructions. The samples were analyzed with a FACSCanto flow cytometer (Beckton Dickinson). At least 10.000 events per sample were recorded. Data were analyzed with FACSDiva software. The median fluorescence values plotted in the graph were background corrected and normalized on CD63/81/9 median signal intensity. Negative values were excluded from the plot.
Statistical analysis
The exact p-value was calculated by un-paired t-test (two groups comparisons) and one-way ANOVA (>2 groups comparisons). Post-hoc t-test values were corrected for multiple comparisons according to Bonferroni correction. The statistical analysis was performed with GraphPad Prism 6 software.
Results
Reduced level of interleukin-6 and DNA damage in cells from centenarians
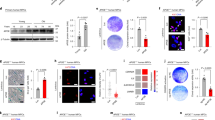
The first observation of our investigation regards the reduced IL-6 protein level in the supernatant of centenarian’s primary DF compared to supernatants from aged people ones (Fig. 1a). The finding above was confirmed by real-time PCR analysis, which also conveyed reduced IL-6 mRNA level in DF and peripheral blood mononuclear cells (PBMC) from centenarians, as compared to aged people (Fig. 1b). These data led us to reason that cells from centenarians are characterized by a restrained pro-inflammatory phenotype. To further confirm this finding, we performed small RNA-seq analysis (Supplementary Figure 1a). The data obtained revealed the down-regulation of a variety of inflammation-related microRNAs (such as miR146a, miR155a, miR21, miR125a, and miR125b) that have been previously associated with age-related diseases [31, 32]. Moreover, anti-inflammatory micro-RNAs such as miR508-3p, miR532-5p, and miR335-5p [33,34,35] were up-regulated in centenarian’s DF (Supplementary Figure 1a). We then tested if such a peculiar molecular set-up persisted even after the induction of a DNA damage: upon exposure to a single radiation dose of 7.5 Gy of gamma rays, we substantiate that centenarian’s DF show a dramatically hampered capability to express IL-6, with respect to cells from old subjects (2 folds vs 80 folds, Fig. 1c). Similar results were observed when DF were exposed to doxorubicin (Fig. 1d). These data indicate that centenarian’s cells are endowed with hampered pro-inflammatory response to DNA damaging agents. This finding is in agreement with our previous observation that centenarian’s DF have a high capability to promptly repair DNA damage [4]. In our set of centenarian’s DF, we found a low level of overall DNA damage measured by comet assay (Fig. 2a). Importantly, such behavior was persistent even after a dose of 7.5 Gy of gamma rays (Fig. 2b). Notably, low levels of the DNA double strand breaks (DSB) sensor Ser139-phosphorylated γ-H2AX were present in centenarian’s DF in basal conditions as well as upon doxorubicin exposure compared to aged people DF (Fig. 2c). Interestingly, we previously reported that centenarian’s DF display a prompt recovery from oxidative stress-induced DNA damage associated with an up-regulation of the DNA damage repair protein 53BP1 [4] which promotes the error-prone NHEJ machinery and switches-off the error-free homologous (HR) DNA repair mechanism [4, 36]. Accordingly, we observed a substantial decrease of the major player of HR protein, i.e., Rad51 in centenarian’s DF compared to old people ones as assessed by western blot and real-time PCR analysis (Fig. 2d). This observation was confirmed by confocal microscopy analysis, showing a decrease of Rad51 positive foci in centenarian’s DF even upon a dose of 7.5 Gy of gamma rays (Fig. 2e). Finally, since telomeres are major source of DNA damage signaling and HR is a major mechanism of telomere maintenance, we assessed telomere length in DF by PCR analysis. In keeping with previous observations [3], we found that centenarian’s DF display higher than expected telomere length, being more similar to young people than old people ones (Fig. 2f). These data show that blunted inflammatory activation in centenarian’s DFs is accompanied by a remarkable genomic stability.
Reduced level of interleukin-6 and high genetic stability in cells from centenarians. a IL-6 ELISA test in aged people (old) and centenarians (cen) DF supernatants. b Real-time PCR analysis of IL-6 mRNA level in old and cen DF (left panel) and peripheral blood mononuclear cells, PBMC (right panel). c IL-6 mRNA level in old and cen DF exposed to gamma rays (7.5 Gy). d IL-6 mRNA level in old and cen DF exposed to doxorubicin (5 µM for 24 h); data are presented as mean ± s.d.
Reduced level of DNA damage in cells from centenarians. a Comet assay in young (yng), aged (old), and centenarians (cen) DF under basal condition. b Comet assay in yng, old, and cen DF upon 7.5 Gy of gamma radiation exposure. c Phospho-serine139-γ-H2AX (γ-H2AX) and actin protein level in yng, old, and cen DF treated or untreated with 5 µM doxorubicin for 24 h. d Rad51, γ-H2AX, and actin protein level in DF from yng, old, and cen, and Rad51 mRNA level in old and cen DF. e Confocal microscopy analysis of Rad51 in yng, old, cen DF at basal condition and upon exposure of 7.5 Gy of gamma radiation. f Telomere/single copy gene ratio (T/S) in DNA from yng (n = 8), old (n = 4), cen (n = 5) DF; data are presented as mean ± s.d.
High level of RNAseH2C and low level of RNA:DNA hybrids in cells from centenarians
DNA damage is a powerful trigger of type I interferon signaling [6, 37]. In this regard, DF from centenarians showed a reduced expression of IFNbeta and IL-6 in comparison to old people cells, while the levels of these cytokines in DF from young subjects were remarkably similar to those of centenarians (Fig. 3a). Notably, an up-regulation of the IFN-regulated gene TREX1 was observed in aged people compared to young and centenarian ones (Fig. 3b). DNA damage elicits inflammation and type I interferon by displacing nucleic acids into the cytoplasm [6, 38]. Cytoplasmic RNA:DNA hybrids are likely to play a substantial role in the above mechanism [19, 20]. These molecules are mainly processed by RNAseH2 enzyme [8, 21]. Remarkably, all three components of the RNAseH2 enzymatic complex were up-regulated in centenarian’s DF compared to old people (Fig. 3c). In particular, the up-regulation of RNAseH2C enzyme subunit in centenarian’s cells was confirmed by western blot analysis (Fig. 3c). Accordingly, the amount of RNA:DNA hybrids, assessed by confocal microscopy analysis, were reduced in the cytoplasm of centenarian’s DF, with respect to old people’s ones (Fig. 3d). Moreover, after exposure to a dose of 7.5 Gy of gamma rays, a dramatic increase in the amount of RNA:DNA hybrids was observed in the cytoplasm of young and old people’s DF, but not in centenarian’s ones (Fig. 3d). Importantly, we observed an opposite relationship between RNAseH2C and IL-6 expression in cells exposed to a dose of 7.5 Gy of gamma rays (Fig. 3e, f). In centenarian’s cells, the transient knock-down of RNAseH2C led to a marked dose-dependent increase of IL-6 and INFbeta expression (Fig. 3g). These data indicate that the anti-inflammatory phenotype of centenarian’s cells is associated with RNAseH2C over-expression.
High level of RNAseH2C and low level of RNA:DNA hybrids in cells from centenarians. a IFN and IL-6 mRNA level in DF from young (yng), aged (old), and centenarian (cen) in a different set of samples respect to Fig. 1b. b TREX1 mRNA level in yng, old, and cen DF. c RNAseH2A-H2B-H2C mRNA level in yng, old, and cen DF; RNAseH2C and actin protein level in yng, old, and cen DF. d Confocal microscopy analysis of RNA:DNA hybrids (S9.6 antibody) in yng, old, and cen DF under basal or upon 7.5 Gy gamma radiation exposure. e RNASeH2C mRNA level in yng, old, and cen DF under basal or upon 7.5 Gy gamma radiation exposure. f IL-6 mRNA level in yng, old, and cen DF under basal or upon 7.5 Gy gamma radiation exposure. g IL-6, IFN, and RNAseH2C mRNA level in cen DF upon RNAseH2C knock-down (20 and 60 ng siRNA); data are presented as mean ± s.d.
RNAseH2C locus methylation in senescent cells and human pathologic tissues
Prompted by the above-described results, we measured RNAseH2C expression in senescent cells [5]. In this latter, an activation of inflammatory and type I interferon signaling due to the misplacement of nucleic acids into the cytoplasm has been recently found [10, 11]. A down-regulation of RNAseH2 subunits expression and a significant up-regulation of IFNbeta were also observed in senescent primary human umbilical endothelial cells (HUVEC) compared to non-senescent ones (Fig. 4a). Accordingly, reduced RNAseH2C expression in senescent fetal lung fibroblasts [26] was found in comparison to early passage cells (Fig. 4b). Epigenetic age-related changes are deeply involved in the aging process [38]. We thus measured DNA methylation at RNAseH2C locus by EpiTYPER (Fig. 4c). A marked hypo-methylation of the first 3 CpG sites of the investigated amplicon was observed in centenarian’s DF (Fig. 4d). In PBMC, we also observed an age-related decrease in the methylation level of RNAseH2C locus at cg11637721 site, which corresponds to the CpG3 site in the EpiTYPER assay (Fig. 4e). Intriguingly, senescent lung fibroblasts displayed a higher degree of methylation at the same RNAseH2C CpG sites that were hypo-methylated in centenarian’s DF (Fig. 4f). We thus reasoned that RNAseH2C gene methylation may be increased in pathologic tissues. To verify this assumption, we searched Gene Expression Omnibus (GEO) database for DNA methylation datasets and analyzed data from GSE46394 and GSE66695 (see Materials and methods). The analysis conveyed that the RNAseH2C CpG site cg11637721 is hyper-methylated in tissues obtained from atherosclerotic plaques (Fig. 4g) and breast cancer tissues, compared to cognate healthy ones (Fig. 4h). These data indicate that disease-free longevity is associated with an un-methylated state of RNAseH2C locus.
Opposite correlation of RNAseH2C locus methylation with longevity and age-related diseases. a RNAseH2C and IFN mRNA level in early passage and senescent HUVEC cells. b RNAseH2C mRNA level in early and senescent (sen) lung fibroblasts. c UCSC view (human GRCh37/hg19) of the RNaseH2C CpG locus explored with EpiTYPER assay and Illumina Infinium 450K corresponding probes. d Percent of methylation for each RNAseH2C CpG unit, measured in young (yng), aged (old), and centenarian (cen) DF. e Percent of methylation of the Illumina Infinium 450K CpG3 unit probe in whole-blood of people of different ages. f Percent of methylation for each RNAseH2C CpG unit, in early and sen human embryonic diploid lung fibroblasts. g Percent of methylation of the Illumina Infinium 450K CpG3 unit probe in normal aortic, aortic, and carotid atherosclerotic samples (dataset GSE46394). h Box plot analysis of percent methylation of CpG3 unit probe in normal and ER-positive or ER-negative breast cancer samples (dataset GSE66695); data are presented as mean ± s.d.
Anti-inflammatory activity of extracellular vesicles (EV) from centenarians
EV have been recently reported to modulate inflammatory and type I interferon signaling pathways [23, 39]. To test the role of EV in our experimental setting, CD9+ EV were isolated from young, old, and centenarian’s DF supernatants, counted by Nanosight and phenotypically characterized by FACS analysis (Supplementary Figure 2a). EV were then administered to primary DF of people of different ages, THP1 myelo-monocytic and MCF-7 breast cancer cells (Fig. 5a). We found that EV from centenarians reduced IL-6 expression and induced RNAseH2C in young people’s DF, as well in THP1 and MCF-7 cells (Fig. 5b, c, Supplementary Figure 2b). In THP1 cells exposed to centenarian’s EV, we observed an over-expression of M2-polarization markers (CD68, CD163, IL-10, and PPAR-gamma) and an up-regulation of anti-inflammatory enzyme (Alox-15) and RXRalpha transcription factor (Fig. 5d, e). A parallel down-regulation of IFNbeta, of the pro-inflammatory cytokine IL-1beta and of PPR-alpha transcription factor were also observed (Fig. 5f). Such an anti-inflammatory capacity of centenarian’s EV was confirmed by the high levels of pro-resolving mediators (maresin-1 and resolvin D1) in THP1 cells administered with centenarian’s EV and exposed to a dose of 7.5 Gy of gamma rays (Fig. 5g). The telomere has been previously characterized among the DNA sequences endowed with peculiar anti-inflammatory activity [40,41,42]. Interestingly, we observed that EV from centenarians contained more than expected amounts of TTAGGG telomeric repeats (Fig. 5h). These data point at the capability of centenarians EV to exert anti-inflammatory activity in recipient cells.
Anti-inflammatory activity of extracellular vesicles from centenarians. a Schematic of the experimental design showing CD9+ EV isolated from young (yng), aged (old), and centenarian (cen) DF and administered to DF, THP1 myelomonocitic cells, and MCF-7 breast cancer cell lines (See Supplementary Figure 2). b IL-6 and RNAseH2C mRNA level in DF exposed to yng-EV, old-EV, cen-EV. c IL-6 and RNAseH2C mRNA level in THP1 cells exposed to yng-EV, old-EV, cen-EV. d IL-10, CD163, CD68, and PPARgamma mRNA level in THP1 cells administered with yng-EV, old-EV, cen-EV. e Alox-15 and RXR-alpha mRNA level in THP1 cells exposed to yng-EV, old-EV, cen-EV. f IFN, IL-1beta, and PPARalpha mRNA level in THP1 cells administered with yng-EV, old-EV, cen-EV. g ELISA test of Maresin1 and Resolvin D1 on the supernatant of THP1 cells administered with yng-EV, old-EV, cen-EV under basal or upon 7.5 Gy gamma radiation exposure. h Real-time PCR analysis of telomeric repeat (T/S) in EV isolated from of yng, old, cen DF; data are presented as mean ± s.d.
Discussion
This paper reports an anti-inflammatory molecular make-up in DF from centenarians. Indeed, as compared to aged people DF, such cells disclose low levels of the pro-inflammatory cytokine IL-6, low expression of INFbeta, as well as an anti-inflammatory microRNA pattern. In particular, microRNA associated with inflammatory age-related diseases such as miR146b, mir21, miR125a/b, miR155a [31, 32] are down-regulated, whereas anti-inflammatory microRNA such as miR508-3p, miR532-5p, and miR335-5p [33,34,35] are up-regulated. These data suggest that the molecular set-up of centenarians may allow these individuals to avoid or postpone inflamm-aging, i.e., the seemingly unavoidable activation of the inflammatory response that occurs during life [2]. Such exceptional individuals are the living proof that it is possible to reach the extreme limits of human lifespan by escaping the detrimental effects of major age-related dysfunctions and diseases [1]. These latter (e.g., atherosclerosis, diabetes, and cancer), albeit extremely heterogeneous, share an inflammatory pathogenesis [12]. Along the myriads of mechanism that may ignite inflammation, DNA damage response has emerged as a crucial trigger of pro-inflammatory/INF alpha/beta response [8, 20, 21, 43]. DNA damage is associated with inflammation in a variety of physiologic and pathologic conditions [6, 8, 37, 43, 44]. Here we show a remarkable low level of DNA damage in cells from centenarians, i.e., almost absent comet tails and barely detectable levels of γ-H2AX phosphorylation, even after exposure to DNA damaging agents. These peculiar features associate with a well-preserved telomere length. All these evidence extend and confirm previous reports [3, 4], showing that centenarians are likely to be endowed with the peculiar capability to repair DNA damage, an activity that is associated with longevity across evolution [45]. Based on the tight relationship between inflammation and DNA damage, the maintenance of the genome is expected to shelter the individuals from a variety of inflammatory diseases, and to prevent or post-pone inflamm-aging. Recent literature shows that DNA damage-induced inflammation and type I interferon signaling depend upon the displacement of nucleic acids into the cytoplasm, where they engage cognate sensors [6, 8, 13, 37]. The phenomenon has been recently linked to the induction of the pro-inflammatory secretome of senescent cells [10]. A particular kind of DNA damage-induced misplaced nucleic acids are RNA:DNA hybrids [6, 19]. Here we show that cells from centenarians are almost devoid of cytoplasmic RNA:DNA hybrids even upon the exposure to gamma radiations. Accordingly, the enzyme involved in the clearance of RNA:DNA hybrids, namely RNAseH2, is strongly up-regulated in cells from centenarians. Then we report that the knock-down of RNAseH2C elicits IL-6 and IFNbeta expression, thus demonstrating that the up-regulation of this enzyme plays a role in the anti-inflammatory set-up of centenarian’s cells. These data are in agreement with current literature showing that RNAseH2C mutations that impair cytoplasmic RNA:DNA hybrids degradation are major trigger for the inflammatory/IFN response [8]. Notably, RNA:DNA hybrids are essential intermediates of the HR-dependent repair of telomere ends but must be proficiently removed to ensure genomic and telomeric stability [46]. Noteworthy, cells of centenarians display almost undetectable levels of Rad51 protein, the major player of HR, thus suggesting that they may act as HR-deficient cells. This apparently paradoxical finding, perfectly fits with our previous observation that centenarian’s cells show the up-regulation of 53BP1, the major mediator of NHEJ, that switches off Rad51 expression and function [36]. In such a peculiar genetic background, the up-regulation of the RNAseH2 enzyme, by promoting RNA:DNA hybrid degradation, is likely assuring genomic stability and telomere maintenance [47]. In this regard, the unbalancing of DNA-repair mechanisms towards NHEJ, associated with higher expression of RNAseH2, is expected to be advantageous for human longevity as it may ensure adequate DNA repair, while avoiding the deleterious effects of Rad51-generated RNA:DNA hybrids [48]. As far as the Rad51 down-regulation in centenarian’s cells, based on our previous observations [26], we observed an up-regulation of Rad51 repressor SLUG, whose down-regulation up-regulated Rad51 mRNA level (Supplementary Figure 3a-b). Noteworthy, SLUG expression was also capable to up-regulate RNAseH2C expression (Supplementary Figure 3c). Though these data may be considered preliminary, it may be speculated that SLUG-dependent mechanisms may control DNA stability and the cognate inflammatory response in aging by modulating RNAseH2 expression. In this regard, we provide evidence that the expression of RNAseH2C is epigenetically regulated. In particular, RNAseH2C is hypo-methylated in centenarians, but hyper-methylated in atherosclerotic plaques and breast cancer tissues. Notably, all these cell types represent pathologic conditions associated with aging and with the up-regulation of inflammatory response [12, 48]. Importantly, our data on the positive relationship between RNAseH2C and longevity are reinforced by literature data showing that RNAseH-dependent RNA:DNA hybrids clearance extends lifespan in the yeast [49]. Moreover, RNAseH2C locus (as well as IL-6 receptor) were identified in a recent genome-wide analysis on human longevity as potential polymorphisms that may explain part of the inter-individual variability in attaining healthy aging and longevity [50]. A major conundrum in aging studies is the relationship between the mechanisms of cellular (in vitro) senescence and those occurring in vivo, at cell and systemic level [14]. In this regard, the accrual of senescent cells, albeit it sets up a minor pool even in aged tissues, has been reported to be sufficient to elicit progeric features in vivo [13]. In particular, the ablation of senescent cells in genetically-engineered mice or via the administration of a set of purposely-named drugs (i.e., senolytics) halts the onset of aging features in vivo [14, 51, 52]. Accordingly, the transplant of few senescent cells into adult mice causes tissue dysfunctions that can be reversed upon the administration of senolytics [14]. As far as the relationship between in vitro senescence in vivo aging is concerned, here we found that in vitro senescent cells convey low expression of RNAseH2C and high RNAseH2C locus methylation. This molecular set-up is expected to facilitate the accumulation in the cytoplasm of nucleic acids (RNA:DNA hybrids in this case) that activate inflammatory/type I interferon signaling in these cells [10, 11, 39]. Nevertheless, cells from centenarians show high levels of RNAseH2C expression and low level of RNAseH2C locus methylation and of RNA:DNA hybrid in the cytoplasm. This molecular make-up seems to favor longevity, at least in part by dampening inflammatory/type I interferon signaling in such cells. In regard to the relationship between cellular and systemic aging features, in this paper we show that centenarian’s cells can cast anti-inflammatory signals to other cell types, such as fibroblasts, monocytes, and cancer cells, via EV. Such subcellular nano-scaled structures have been extensively characterized [24] and contain a wealth of cellular components, including nucleic acids [34]. In all cellular models here tested, EV isolated from centenarian’s DF are capable to induce RNAseH2C and reduce IL-6 expression. EV from centenarians also skew monocyte polarization towards the M2 phenotype which exhibits anti-inflammatory, pro-resolution, and reparative capability [53]. These data are reinforced by our observation regarding the capability of EV from centenarians to up-regulate two omega 3–derived lipid mediators, Resolvin-D1 and Maresin, which are tightly linked to the M2 macrophage phenotype [54, 55]. These anti-inflammatory mediators are currently regarded as molecules with a powerful capability to taper inflammation and to promote tissue integrity and repair [55]. Interestingly, EV from centenarians administered to MCF-7 grown in vitro as 3D cultures (mammospheres, MS) [54], blunted MS growth and reduced the level of the IL-6 dependent MS growth factor JAG1 (Supplementary Figure 4a and 4b) [54]. These data suggest that EV from long-living individuals may halt or thwart cancer cell growth. The amount of RNA in EV did not allow us to assess whether the microRNA content of such EV may propagate the anti-inflammatory phenotype to target cells. However, as a support of the centenarian’s EV anti-inflammatory activity we could demonstrate the presence of higher than expected telomeric DNA sequences TTAGGG in centenarian’s EV. Indeed, extracellular telomere sequences exert potent inhibitory activity on inflammation/type I interferon signaling [56, 57]. These data suggest that EV may spread the anti-inflammatory set-up of centenarian’s cells at a systemic level, thus providing cues on the molecular mechanisms that taper inflamm-aging and promote disease-free longevity in humans.
References
Franceschi C, Bonafe M. Centenarians as a model for healthy aging. Biochem Soc Trans. 2003;31:457–61.
Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, et al. Inflammaging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–54.
Mondello C, Petropoulou C, Monti D, Gonos ES, Franceschi C, Nuzzo F. Telomere length in fibroblasts and blood cells from healthy centenarians. Exp Cell Res. 1999;248:234–42.
Lattanzi G, Ortolani M, Columbaro M, Prencipe S, Mattioli E, Lanzarini C, et al. Lamins are rapamycin targets that impact human longevity: a study in centenarians. J Cell Sci. 2014;127:147–57.
Rodier F, Coppe JP, Patil CK, Hoeijmakers W, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–79.
Shen YJ, Le Bert N, Chitre AA, Ko CX, Nga XH, Ho SS, et al. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015;11:460–73.
Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–80.
Crow YJ, Manel N. Aicardi–Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–40.
Molès JP, Griez A, Guilhou JJ, Girard C, Nagot N, Van de Perre P, et al. Cytosolic RNA:DNA duplexes generated by endogenous reverse transcriptase activity as autonomous inducers of skin inflammation in psoriasis. PLoS One. 2017;12:e0169879.
Gluck S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19:1061–70.
Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci USA. 2017;114:E4612–20.
Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21:1424–35.
Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69:S4–9.
Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, et al. Senolytics improve physical function and increase lifespan in old age. Nat Med. 2018;24:1246–56.
Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–58.
Ershler WB. Interleukin-6: a cytokine for gerontologists. J Am Geriatr Soc. 1993;41:176–81.
Knupfer H, Preiss R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res Treat. 2007;102:129–35.
Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, et al. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep. 2015;11:785–97.
Koo CX, Kobiyama K, Shen YJ, LeBert N, Ahmad S, Khatoo M, et al. RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J Biol Chem. 2015;290:7463–73.
Mankan AK, Schmidt T, Chauhan D, Goldeck M, Höning K, Gaidt M, et al. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33:2937–46.
Günther C, Kind B, Reijns MA, Berndt N, Martinez-Bueno M, Wolf C, et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clin Invest. 2015;125:413–24.
Keskin H, Storici F. Defects in RNase H2 stimulate DNA break repair by RNA reverse transcribed into cDNA. MicroRNA. 2015;4:109–16.
Kitai Y, Kawasaki T, Sueyoshi T, Kobiyama K, Ishii KJ, Zou J, et al. DNA-containing exosomes derived from cancer cells treated with topotecan activate a STING-dependent pathway and reinforce antitumor immunity. J Immunol. 2017;198:1649–59.
Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–32.
Eitan E, Green J, Bodogai M, Mode NA, Baek R, Jorgensen MM, et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci Rep. 2017;7:1342.
Bacalini MG, Deelen J, Pirazzini C, De Cecco M, Giuliani C, Lanzarini C, et al. Systemic age-associated DNA hypermethylation of ELOVL2 gene: in vivo and in vitro evidences of a cell replication process. J Gerontol A Biol Sci Med Sci. 2017;72:1015–23.
Storci G, Bertoni S, De Carolis S, Papi A, Nati M, Ceccarelli C, et al. Slug/beta-catenin-dependent proinflammatory phenotype in hypoxic breast cancer stem cells. Am J Pathol. 2013;183:1688–97.
Prattichizzo F, Giuliani A, Recchioni R, Bonafe M, Marcheselli F, De Carolis S, et al. Anti-TNF-alpha treatment modulates SASP and SASP-related microRNAs in endothelial cells and in circulating angiogenic cells. Oncotarget. 2016;7:11945–58.
Tesei A, Sarnelli A, Arienti C, Menghi E, Medri L, Gabucci E, et al. In vitro irradiation system for radiobiological experiments. Radiat Oncol. 2013;8:257.
Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47.
Olivieri F, Rippo MR, Procopio AD, Fazioli F. Circulating inflamma-miRs in aging and age-related diseases. Front Genet. 2013;4:121.
Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci USA. 2012;109:7865–70.
Jia-Xing Z, Zhen-Hua C, Dong-Liang C, Xiao-Peng T, Chen-Yuan W, Zhi-Wei Z, et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660–75.
Gao XL, Li JQ, Dong YT, Cheng EJ, Gong JN, Qin YL, et al. Upregulation of microRNA-335-5p reduces inflammatory responses by inhibiting FASN through the activation of AMPK/ULK1 signaling pathway in a septic mouse model. Cytokine. 2018;110:466–78.
Huang T, Kang W, Zhang B, Wu F, Dong Y, Tong JH, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-kappaB signaling in gastric carcinogenesis. Mol Cancer. 2016;15:9.
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204.
Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31:353–69.
Horvath S, Pirazzini C, Bacalini MG, Gentilini D, Di Blasio AM, Delledonne M, et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY). 2015;7:1159–70.
Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287.
Gursel I, Gursel M, Yamada H, Ishii KJ, Takeshita F, Klinman DM. Repetitive elements in mammalian telomeres suppress bacterial DNA-induced immune activation. J Immunol. 2003;171:1393–00.
Shirota H, Gursel I, Gursel M, Klinman DM. Suppressive oligodeoxynucleotides protect mice from lethal endotoxic shock. J Immunol. 2005;174:4579–83.
Goldfarb IT, Adeli S, Berk T, Phillippe M. Fetal and placental DNA stimulation of TLR9: a mechanism possibly contributing to the pro-inflammatory events during parturition. Reprod Sci. 2017;25:788–96.
Brégnard C, Guerra J, Déjardin S, Passalacqua F, Benkirane M, Laguette N. Upregulated LINE-1 activity in the fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine. 2016;8:184–94.
Bonafè M, Storci G, Franceschi C. Inflamm-aging of the stem cell niche: breast cancer as a paradigmatic example: breakdown of the multi-shell cytokine network fuels cancer in aged people. Bioessays. 2012;34:40–9.
MacRae SL, Zhang Q, Lemetre C, Seim I, Calder RB, Hoeijmakers J, et al. Comparative analysis of genome maintenance genes in naked mole rat, mouse, and human. Aging Cell. 2015;14:288–91.
Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. Transient RNA–DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–13.
Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, et al. Telomeric RNA–DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–205.
Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. eLife. 2013;2:e00505.
Salvi JS, Chan JN, Szafranski K, Liu TT, Wu JD, Olsen JB, et al. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA–DNA hybrids. Dev Cell. 2014;30:177–91.
Pilling LC, Kuo CL, Sicinski K, Tamosauskaite J, Kuchel GA, Harries LW, et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging (Albany NY). 2017;9:2504–20.
Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–6.
Baar MP, Brandt RMC, Putavet DA, Klein JDD, Derks KWJ, Bourgeois BRM, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–47.
Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–85.
Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–02.
Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101.
Marwaha V, Chen YH, Helms E, Arad S, Inoue H, Bord E, et al. T-oligo treatment decreases constitutive and UVB-induced COX-2 levels through p53- and NFkappaB-dependent repression of the COX-2 promoter. J Biol Chem. 2005;280:32379–88.
Steinhagen F, Zillinger T, Peukert K, Fox M, Thudium M, Barchet W, et al. Suppressive oligodeoxynucleotides containing TTAGGG motifs inhibit cGAS activation in human monocytes. Eur J Immunol. 2018;48:605–11.
Acknowledgements
The authors acknowledge Prof. John Sedivy (Department of Molecular Biology, Brown University, Providence, RI, USA) for providing DNA of fetal lung fibroblasts; Antonino Romeo (Radiotherapy Unit, IRST-IRCCS Meldola, Forlì) for their technical assistance for gamma radiation exposure experiments; Michela Cortesi and Sara Pignatta (Bioscience Laboratory, IRST-IRCCS Meldola, Forlì) for their technical assistance in comet assay and cytofluorimetric analysis experiments. A particular acknowledge to Michele Guescini (University of Urbino) for his technical assistance on EV isolation and characterization and to Cristina Fabbri (University of Bologna) for primary fibroblasts cultures. This work has been supported by (1) the EU-FP7 Projects under the grant agreements no. 613979 “MyNewGut”; no. 602757 “HUMAN” to CF; (2) by the EU-H2020 Project under the grant agreement no. 634821 “Propag-ageing” to CF; (3) by Roberto and Cornelia Pallotti Legacy for Cancer Research (University of Bologna) to MB and SS; (4) by Bologna AIL (Italian Association against Leukemia, Lymphoma and Myeloma.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Storci, G., De Carolis, S., Papi, A. et al. Genomic stability, anti-inflammatory phenotype, and up-regulation of the RNAseH2 in cells from centenarians. Cell Death Differ 26, 1845–1858 (2019). https://doi.org/10.1038/s41418-018-0255-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0255-8
This article is cited by
-
Pro-inflammatory RNA:DNA Hybrids Are p53 Independently Boosted by Hyperbaric Oxygen: a Subcellular Distribution Analysis by Automated Quantitative Imaging
Molecular Imaging and Biology (2023)
-
Dauer larva-derived extracellular vesicles extend the life of Caenorhabditis elegans
Biogerontology (2023)
-
The multifaceted role of the SASP in atherosclerosis: from mechanisms to therapeutic opportunities
Cell & Bioscience (2022)
-
Aging and aging-related diseases: from molecular mechanisms to interventions and treatments
Signal Transduction and Targeted Therapy (2022)
-
TP53 drives abscopal effect by secretion of senescence-associated molecular signals in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2021)