Abstract
MicroRNAs (miRNAs) play significant roles in both embryonic hematopoiesis and hematological malignancy. Zebrafish miR-462-731 cluster is orthologous of miR-191-425 in human which regulates proliferation and tumorigenesis. In our previous work, miR-462-731 was found highly and ubiquitously expressed during early embryogenesis. In this study, by loss-of-function analysis (morpholino knockdown combined with CRISRP/Cas9 knockout) and mRNA profiling, we suggest that miR-462-731 is required for normal embryonic development by regulating cell survival. We found that loss of miR-462/miR-731 caused a remarkable decrease in the number of erythroid cells as well as an ectopic myeloid cell expansion at 48 hpf, suggesting a skewing of myeloid-erythroid lineage differentiation. Mechanistically, miR-462-731 provides an instructive input for pu.1-dependent primitive myelopoiesis through regulating etsrp/scl signaling combined with a novel pu.1/miR-462-731 feedback loop. On the other hand, morpholino (MO) knockdown of miR-462/miR-731 resulted in an expansion of posterior blood islands at 24 hpf, which is a mild ventralization phenotype resulted from elevation of BMP signaling. Rescue experiments with both BMP type I receptor inhibitor dorsomorphin and alk8 MO indicate that miR-462-731 acts upstream of alk8 within the BMP/Smad signaling pathway and functions as a novel endogenous BMP antagonist. Besides, an impairment of angiogenesis was observed in miR-462/miR-731 morphants. The specification of arteries and veins was also perturbed, as characterized by the irregular patterning of efnb2a and flt4 expression. Our study unveils a previously unrecognized role of miR-462-731 in BMP/Smad signaling mediated hematopoietic specification of mesodermal progenitors and demonstrates a miR-462-731 mediated regulatory mechanism driving primitive myelopoiesis in the ALPM. We also show a requirement for miR-462-731 in regulating arterial-venous specification and definitive hematopoietic stem cell (HSC) production. The current findings might provide further insights into the molecular mechanistic basis of miRNA regulation of embryonic hematopoiesis and hematological malignancy.
Similar content being viewed by others
Introduction
In vertebrates, hematopoiesis occurs in two successive waves during embryonic development [1]. In zebrafish embryos, primitive hematopoiesis originates in the ventral mesoderm, later forming an intermediate cell mass (ICM) [2]. Definitive hematopoiesis occurs within the ventral wall of the dorsal aorta (DA) in the aorta-gonad-mesonephros (AGM) region as evidenced by the expression of c-myb and runx1 [3], then migrates to the caudal hematopoietic tissue (CHT) in the tail, and eventually switches to the kidney marrow, where life-long hematopoiesis occurs [1, 4]. Processes required for hematopoiesis already underway when embryos develop three germ layers, among which blood and angioblasts originate from the mesoderm [5]. Bone morphogenetic proteins (BMPs), growth factors belonging to the transforming growth factor β (TGF-β) superfamily, are essential for dorsoventral axis formation [6]. Recent evidences revealed that activin receptor-like kinase 8 (Alk8), a novel type I TGF-β receptor, is required for Bmp2b/7 signaling mediated dorsoventral patterning [7, 8]. BMP signaling also regulates hematopoietic stem cell (HSC) development during embryonic hematopoiesis [9,10,11].
In zebrafish, hematopoietic and endothelial lineages derive from a common multipotent hematopoietic progenitor, the hemangioblast [12]. The differentiation of hemangioblasts from the ventral lateral mesoderm is regulated by several transcription factors, including etsrp, fli1a, scl, lmo2, and gata2 [13]. Primitive myelopoiesis arises from the anterior lateral plate mesoderm (ALPM), characterized by pu.1 expression [14, 15]. These pu.1+ myeloid progenitors then give rise to granulocytes and macrophages characterized by mpo and l-plastin expression [16, 17]. In parallel, the majority of the hemangioblasts in the posterior lateral plate mesoderm (PLPM) become gata1+ erythroid progenitors which thereafter develop into mature erythrocytes expressing hemoglobin [18]. The interplay between pu.1 and gata1 directs the differentiation of myeloerythroid progenitor cells into myeloid and erythroid lineages [19, 20].
A group of microRNAs (miRNAs) have been shown to be expressed in specific hematopoietic lineages and play significant roles in both hematopoiesis and hematological malignancy [21]. In zebrafish, some miRNAs were suggested to regulate hematopoietic differentiation, myelopoiesis, and vascular development during embryogenesis [22,23,24]. Teleost-conserved miR-462-731 cluster had evolved from the ancestral miR-191-425 in human, which was recently reported to promote tumorgenesis by regulating TGF-β signaling [25, 26]. As a hematopoietic lineage-enriched miRNA [27], miR-191 also regulates erythroblast enucleation, angiogenesis and T cell survival [28,29,30]. Our previous study revealed that zebrafish miR-462-731 regulates cell survival in response to hypoxia and is highly and ubiquitously expressed during early embryogenesis [31]. However, the potential for miR-462-731 regulation of hematopoiesis has not been experimentally addressed, and the mechanism underlying this process remains to be elucidated.
In this study, we provide evidence that miR-462-731 is essential for zebrafish normal embryonic development by regulating cell survival. Furthermore, by loss-of-function analysis and rescue experiments, we suggest that miR-462-731 acts as a molecular determinant for hematopoietic lineage differentiation and vascular specification, and regulates dorsoventral patterning via Alk8-mediated BMP/Smad signaling.
Results
MiR-462-731 regulates normal embryonic development
To systematically define functions of miR-462-731 during embryonic development, two Morpholinos (MOs) that target the mature sequence of each miRNA were microinjected, respectively. Quantitative real-time PCR (qRT-PCR) confirmed that miR-462 and miR-731 were effectively and continuously suppressed from 12 h post-fertilization (hpf) to 5 days post-fertilization (dpf) (Supplementary Figure S1A). MO-injected embryos showed a developmental defect with curved tail, small eyes, and pericardial edema; they were incapable of forming swim bladder (Fig. 1a). The morphologic abnormality was observed in a dose-dependent manner (Supplementary Figure S1B), with 4–8 ng miR-462 MO and 1–2 ng miR-731 MO can reproducibly achieve a malformation rate of >50% (Supplementary Figure S1C). To avoid potential off-targeting effects by using high concentrations, we used 4 ng and 1 ng for miR-462 and miR-731 MO, respectively, in all subsequent experiments. Careful examination revealed that blood circulation and red blood cells were remarkably reduced or absent in miR-462/miR-731 morphants (Fig. 1a). And this phenotype may be partially rescued by corresponding miRNA duplex in a dose-dependent manner (Supplementary Figure S1D).
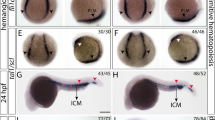
MiR-462/miR-731 knockdown disrupts normal embryonic development. a Injection of miR-462/miR-731 MOs induces distinct developmental defects from 48 hpf to 5dpf. sb, swim bladder; nsb, non-swim bladder; pe, pericardial edema. b Detection of cell death in ICM region at 24 hpf, head and tail at 48 hpf and pericardium at 72 hpf by AO staining. c Apoptosis phenotype in the tail verified by TUNEL assay. Scale bars, 0.2 mm
To further confirm the specificity of MO phenotypes, knockout of miR-462-731 cluster was performed by CRISPR/Cas9. Two target sites were designed to delete a 563-bp fragment holding the miR-462-731 cluster, which finally resulted in a deletion of a 579-bp fragment containing both pre-miRNAs (Supplementary Figure S2A). Two pairs of primers were designed for genotyping. When using the OuterPrimers, a specific band of 392-bp was detected in heterozygote and homozygote, whereas only a 971-bp full-length fragment was observed in wild type. When using the InnerPrimers, a 224-bp band was detected in wild type and heterozygote, but not in homozygote (Supplementary Figure S2B). Around 30% of the F2 were identified as homozygote, and these embryos developed with the same defects observed in miR-462/miR-731 morphants at 48 hpf (Supplementary Figure S2C).
By conducting mRNA profiling in miR-462 and miR-731 morphants at 48 hpf, we identified a total of 863 and 744 differentially expressed genes (DEGs), respectively, with a notable overlap (Supplementary Figure S3, Supplementary Tables S1 and S2). Gene ontology (GO) enrichment revealed that developmental process (GO:0032502) was significantly affected (P462 = 5.80E−08, P731 = 8.73E−06), especially with upregulated genes enriched in muscle development and downregulated genes annotated to the nervous system, brain, eye development, and hemoglobin complex (Supplementary Figure S4). Notably, p53 signaling and glycolysis were significantly upregulated, but Wnt signaling and phagosome were downregulated (Supplementary Table S3). DEGs implicated in Wnt and p53 signaling were further clustered (Supplementary Figure S5A). As verified by qRT-PCR, expression of lef1 and tcf7l2 was decreased, while genes responsible for cell cycle arrest such as cdkn1a (p21) and gadd45aa, and factors promoting proliferation including mycb and fols1a were dramatically upregulated (Supplementary Figures S5B and C). We further identified p21 as a bona fide target of miR-462 (Supplementary Figure S6). Meanwhile, apoptosis regulators casp8 and casp3a were significantly induced and revealed to be negatively regulated by miR-462 and miR-731, respectively (Supplementary Figure S7). Whole-mount Acridine Orange (AO) staining and terminal transferase-mediated deoxyuridine triphosphate nick end-labelling (TUNEL) assay revealed that miR-462/miR-731 knockdown resulted in significant apoptosis in brain, eye, and tail at 48 hpf and in pericardium at 72 hpf (Fig. 1b, c). This result was further confirmed in knockout homozygote (Supplementary Figure S8), suggesting that miR-462-731 specifically affects normal embryonic development by regulating cell survival.
MiR-462-731 regulates erythropoiesis
Among the DEGs, a set of genes related to hematopoiesis, circulatory system, and blood vessel development were dysregulated (Fig. 2a). Among them, genes encoding α/β subunits of hemoglobin (hbae1, hbae3, hbbe1, and hbbe2), alas2, and fli1a were confirmed to be significantly downregulated in MO-injected embryos (Fig. 2b). We further verified the hematopoietic defects observed in visual inspection by o-dianisidine staining. From 36 hpf to 96 hpf, depletion of miR-462/miR-731 caused a remarkable decrease in the number of erythroid cells as well as blood flow obstruction (Fig. 2c). The difference was persistent and became pronounced during the development, indicating that the phenotype was not simply due to developmental delay. And this result was further verified in miR-462-731 knockout homozygous embryos (Supplementary Figure S9). Furthermore, a significant decrease in hemoglobin expression and blood circulation was observed in whole-mount in situ hybridization (WISH) detection of hbbe2, hbae1, and alas2 at 48 hpf, which can be partially rescued by corresponding miRNA duplex (Fig. 2d). These results indicate that miR-462/miR-731 knockdown specifically affects erythropoiesis during embryonic development.
Erythropoiesis is defective in miR-462/miR-731 morphants. a Clustering analysis of differentially expressed genes related to hematopoiesis, circulatory system and blood vessel development in control and miR-462/miR-731 morphants at 48 hpf. b QRT-PCR verification of the expression of hemoglobin, alas2 and fli1a. Expression is normalized to 18s rRNA. Error bars represent SD (N = 3). *P < 0.05, **P < 0.01. c O-dianisidine staining for hemoglobin in MO-injected embryos from 36 hpf to 96 hpf. d WISH for hemoglobin at 48 hpf. Arrow indicates reduced erythroid cells and blood circulation. Scale bars, 0.2 mm. WISH detection was performed for two times
Loss of miR-462/miR-731 induces ectopic myeloid cell expansion
We consider further effect on myeloid cell development. On the contrary, WISH and qRT-PCR results revealed that expression of both l-plastin and mpo were massively induced from 24 hpf to 48 hpf (Fig. 3a, Supplementary Figure S10A and B). MO knockdown was further performed in Tg(mpo:GFP), Tg(lyz:GFP) and Tg(coro1a:GFP). Similarly, the population of mpo-GFP+ and lyz-GFP+ neutrophils, and coro1a-GFP+ leukocytes was increased significantly at 48 hpf (Fig. 3b). And the induction of neutrophils maintained and became pronounced at 72 hpf (Supplementary Figure S10C). This ectopic expansion of myeloid cells can be partially rescued by corresponding miRNA duplex. These results suggest a skewing of myeloid-erythroid lineage differentiation in miR-462/miR-731 morphants.
MiR-462/miR-731 morphants display an induction of primitive myelopoiesis
We then performed WISH for key hematopoietic transcription factors in the ventral lateral mesoderm to test whether miR-462-731 regulates primitive hematopoiesis. It was shown that miR-462/miR-731 knockdown significantly increased the expression of etsrp and scl, primarily in the ALPM, and affected both the amount of expression and size of the area (Fig. 4a, Supplementary Figure S11A). In the PLPM, scl, etsrp, and lmo2 were also upregulated slightly. But, fli1a was quite unaffected (Fig. 4a). Here, lmo2 was also experimentally demonstrated as a bona fide target of miR-731 (Supplementary Figure S6). We further investigated whether miR-462/miR-731 knockdown affects the expression of nonhemangioblast-related mesoderm markers by performing WISH for myod (muscle), ntl (notochord), pax2a (pronephros) and nkx2.5 (cardiomyocytes). The only nkx2.5 level was slightly decreased, the expression of other markers was not apparently changed (Supplementary Figure S12), indicating that miR-462-731 specifically regulates hemangioblast specification and hematopoietic lineage differentiation.
MiR-462-731 regulates hematopoietic specification and ALPM myelopoiesis. a WISH for hemangioblast markers scl, lmo2, etsrp, and fli1a at 10-somite stage. b WISH for the myeloid progenitor marker pu.1 at 10-somite stage and 24 hpf. Arrow indicates induction of pu.1 in the head and ICM region. c WISH for the erythroid progenitor marker gata1 at 10-somite stage and 24 hpf. Scale bars, 0.2 mm. Arrow indicates posterior ICM expansion. WISH detection was performed for three times
Along with the induction of scl, myeloid progenitor specific pu.1 expression was severely increased in the ALPM at 10-somite stage and the induction remained at 24 hpf both in the anterior part and the posterior ICM (Fig. 4b, Supplementary Figure S11B). Recently, alk8 signaling was reported to function in parallel to the scl signaling in pu.1-dependent ALPM myelopoiesis [32]. We tested if miR-462/miR-731 knockdown could induce ectopic pu.1 expression in alk8 morphants. As expected, miR-462 knockdown failed to rescue pu.1 expression in the ALPM in alk8 morphants (Fig. 4b). But, alk8 is not required for miR-731 MO caused pu.1 induction (Fig. 4b), implying a more direct regulation of pu.1 by miR-731. On the other hand, the expression of gata1 was reduced in the PLPM at 10-somite stage and the loss of gata1 transcript was still evident at 24 hpf in the ICM (Fig. 4c, Supplementary Figure S11C), further suggesting that miR-462/miR-731 knockdown not only upregulated pu.1 expression, but also disrupted the interplay between pu.1 and gata1.
MiR-731 negatively regulates pu.1 which in turn induces miR-462-731 expression
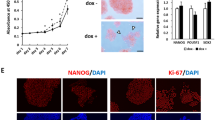
A Pu.1 responsive element in the promoter region of miR-462-731 was found highly conserved among teleosts [33]. We therefore investigated the transcriptional regulation of miR-462-731 by Pu.1. Two putative Pu.1 responsive elements were identified on the promoter of zebrafish miR-462-731 (Fig. 5a). Western blot analysis confirmed the overexpression of zebrafish Pu.1 in HeLa cells by transfecting pCMV-Myc-Pu.1 construct (Fig. 5b). Luciferase reporter assay showed that the wildtype promoter fragment is highly responsive to Pu.1 activation, while mutation of site 1 resulted in a significant decrease of reporter activity, suggesting that site 1 can be effectively targeted by Pu.1. However, site 2 is noneffective because no significant difference of luciferase activity was observed in site 2-mutant (Fig. 5c). Moreover, endogenous pu.1 expression was effectively suppressed by siRNA transfection in ZF4 cells (Fig. 5d), and the expression level of both miR-462/miR-731 and primary transcript was significantly decreased accordingly (Fig. 5e). On the contrary, the expression of miR-462-731 was upregulated in the pu.1-overexpressing embryos (Fig. 5f, g).
MiR-731 negatively regulates pu.1 which in turn induces miR-462-731 expression. a A schematic depiction of the miR-462-731 promoter region containing two putative Pu.1 binding sites. b Protein level detection of recombinant zebrafish Myc-tagged-Pu.1 (37.5 kDa) in HeLa cells. The expression is normalized to β-actin. c Luciferase reporter assay for four indicated promoter constructs. pCMV-myc is used as control. Firefly luciferase expression was normalized to Renilla luciferase (internal control). d QRT-PCR detection for endogenous expression of pu.1 in ZF4 cells transfected with Pu.1 siRNA (si-NC, negative control siRNA). e Quantification of miR-462/miR-731 and primary transcript (pri-miR) in ZF4 cells transfected with Pu.1 siRNA based on qRT-PCR. f QRT-PCR detection for endogenous expression of pu.1 in embryos injected with pu.1 mRNA. g Quantification of miR-462/miR-731 and primary transcript (pri-miR) in in embryos injected with pu.1 mRNA based on qRT-PCR. (h) Schematic illustration of pu.1 3’UTR fragment harboring a miR-731 binding site. (i) Dual-luciferase reporter assay for validation of miR-731 binding site on the pu.1 3’UTR. Renilla luciferase activities were detected and normalized to Firefly luciferase (internal control). j QRT-PCR detection for injection efficiency of miR-731 duplex and MO in zebrafish embryos. k Negative regulation of pu.1 expression by miR-731 detected by qRT-PCR. All relative miRNA expression is normalized to U6-1, and mRNA expression is normalized to 18s rRNA. All results are presented as mean ± SD (N = 3), *P < 0.05, **P < 0.01, NS, not significant
On the other hand, pu.1 was identified as a potential target of miR-731, with a complementary binding site on the 3’-UTR (Fig. 5h). Overexpression of miR-731 significantly reduced the relative luciferase activity. The specificity of this inhibition was evidenced by the finding that the activity of a mutant 3’-UTR construct was not affected (Fig. 5i). Knockdown and overexpression were conducted by injecting 731 MO or duplex in zebrafish embryos (Fig. 5j). Endogenous pu.1 expression was considerably increased by miR-731 MO, but decreased by miR-731 duplex (Fig. 5k). These results characterize a previously unrecognized pu.1/miR-462-731 negative regulatory feedback loop which may play a role in hematopoietic lineage differentiation.
MiR-462-731 regulates Alk8-mediated BMP/Smad signaling
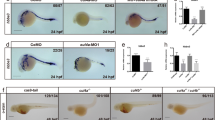
Notably, at 24 hpf, the onset of blood cell circulation, we observed an obvious expansion of posterior ICM area (Fig. 4c, arrow). Careful examination revealed a slight reduction of the head and an expansion of ventroposterior blood islands in the miR-462/miR-731 morphants (Fig. 6a). Closer investigation indicated a significant increase in the size of the posterior ICM area (Fig. 6a). This weakly ventralized phenotype (class II) may result from an elevation of BMP signaling [34, 35]. Therefore, we detected the expression of phosphorylated (P)-Smad1/5 and alk8, and revealed that knockdown of miR-462/miR-731 caused a pronounced induction in P-Smad1/5 levels (Fig. 6b) accompanied by an increase of alk8 expression during gastrulation (Fig. 6c). Co-injection of alk8 MO rescued the ectopic expansion of hematopoietic mesoderm (Fig. 6a). We further investigated the hierarchical relationship of miR-462-731 with dorsomorphin, a small-molecule BMP antagonist that selectively inhibits BMP type I receptors Alk2 (Alk8), Alk3 and Alk6 [36]. Dorsomorphin treatment reversed the moderate ventralization phenotype caused by loss of miR-462/miR-731 (Fig. 6a).
Loss of miR-462/miR-731 caused mild ventralization phenotype and elevated BMP/Smad signaling. a Morphological defects in miR-462/miR-731 morphants at 24 hpf. MiR-462/miR-731 morphants showed enlarged posterior ICM (region marked in red), which can be rescued by co-treatment with dorsomorphin (DM) or co-injection with alk8 MO. Scale bars, 0.2 mm. b Western blot analysis for phosphor-Smad1/5 levels in control embryos and miR-462/miR-731 morphants at 70%-epiboly stage. Expression is normalized to β-actin. c Quantification of alk8 expression at 70%-epiboly stage by qRT-PCR. Expression is normalized to 18 s rRNA. Error bars represent SD (N = 3). *P < 0.05, **P < 0.01. WISH detection was performed for three times
Knockdown of miR-462/miR-731 perturbs vascular development and definitive HSC production
We further examined whether the observed defects in blood flow could result from perturbation of vasculogenesis. By WISH detection of fli1a at 24 hpf, we observed an obvious expansion of posterior ICM along with significant caudal vascular disturbance (Fig. 7a). Angiogenesis, as shown by sprouting of intersegmental vessels (ISVs), was significantly perturbed in MO-injected embryos with a Tg(fli1a:GFP) transgenic background (Fig. 7a, Supplementary Figure S13). At 48 hpf, miR-462/miR-731 knockdown resulted in DA and cardinal vein (CV) abnormality, and loss of dorsal longitudinal anastomotic vessel (DLAV) integrity. The ISVs sprout and extend dorsally but are irregularly patterned and fail to make contact with DLAV (Fig. 7b, Supplementary Figure S13). The defect in angiogenesis was persistent at 72 hpf, indicating that the phenotype was not simply due to developmental delay. Additionally, the morphants displayed dramatic reduction of efnb2a (arterial marker) and ectopic expansion of flt4 (venous marker) in the posterior cardinal vein (PCV), which is in consistent with the irregular patterning of flk1 expression at 36 hpf (Fig. 7c, Supplementary Figure S11D). The axial blood circulation was also severely disrupted as shown in MO-injected embryos with a Tg(flk1:GFP/gata1:DsRed) transgenic background at 48 hpf (Fig. 7d, arrow). These data demonstrate that loss of miR-462/miR-731 perturbed both angiogenic sprouting and arterial/venous programming, leading to circulation defect.
Knockdown of miR-462/miR-731 perturbs vascular development and definitive hematopoiesis. a WISH for fli1a and confocal images of Tg(fli1a:GFP) at 24 hpf. Black arrow indicates posterior ICM expansion; White arrow indicates intersegmental vessels (ISVs) defects. b WISH for fli1a and confocal images of Tg(fli1a:GFP) in the trunk region at 48 hpf. Red arrow indicates dorsal aorta (DA); Blue arrow indicates cardinal vein (CV); yellow arrow indicates ISV. c WISH for flk1, flt4, and efnb2a at 36 hpf. d Confocal images of Tg(flk1:GFP/gata1:DsRed) at 48 hpf show a decreased number of erythroid cells in circulation (white arrow) in miR-462/miR-731 morphants. Arrow indicates disruption of axial blood circulation. e WISH for c-myb at 36 hpf and rag1 at 96 hpf. Arrowheads and the arrow indicate c-myb (AGM region) and rag1 (thymus) expression, respectively. Scale bars, 0.2 mm. WISH detection was performed for two times
The definitive HSCs emerge in the ventral wall of the DA, so we further examined the effect on definitive hematopoiesis. The number of c-myb-expressing HSCs was significantly reduced in the ventral wall of DA in miR-462/miR-731 morphants at 36 hpf (Fig. 7e, arrowhead). Also, the expression of rag1 in the thymus was dramatically downregulated and almost undetectable at 96 hpf (Fig. 7e, arrow). These results were confirmed by qRT-PCR (Supplementary Figure S11E).
Discussion
Zebrafish miR-462-731 has been reported ubiquitously expressed during early embryogenesis, with strong signal detected in eyes, brain, and somatic muscle [31]. Our analysis of morphologic abnormality combined with GO enrichment of DEG sets suggests that miR-462-731 has an important requirement in the developmental process. And a dysregulation of the canonical Wnt/β-Catenin and p53 signaling may be responsible for the defects in embryonic development and organ formation [37, 38]. Teleost-conserved miR-462-731 had evolved from the ancestral miR-191-425 in human, which is involved in proliferation and tumorigenesis [39, 40]. Evidences have demonstrated a pivotal role for miR-462-731 in regulating apoptosis and cell cycle arrest via p53 signaling [31, 41]. Here, significant apoptosis signal was detected in brain, eye, and trunk in both MO-injected embryos and miR-462-731 knockout homozygote, meanwhile genes involved in cell cycle arrest (p21) and apoptosis (casp8 and casp3a) were identified as novel targets. These results support a crucial role for miR-462-731 during embryogenesis through regulating cell survival.
Besides, both miR-462/miR-731 morphants and miR-462-731-null mutant developed with a striking deficit of erythroid cell and blood circulation at 48hpf. Meanwhile, a loss of hemoglobin expression was detected, along with downregulation of a set of hematopoiesis-related genes, including jak2a, pbx1, lef1, and fli1a. Observed hematopoietic defects may not be associated with p53-incuced apoptosis, since no increase of apoptosis signal in the ICM at 24 hpf or along the ventral wall of DA at 48 hpf was observed. And we further observed an ectopic expansion of myeloid cells, suggesting a skewing of myeloid-erythroid lineage differentiation.
During the primitive hematopoiesis, loss of miR-462/miR-731 caused an expansion of hemangioblasts, characterized by a dramatic induction of hematopoietic transcription factors. Among them, scl and lmo2 act in parallel to maintain the hemangioblast population and regulate differentiation of the hemangioblast into erythrocytes and myeloid cells [42]. Our results established miR-462-731 as a molecular determinant for primitive myelopoiesis. MiR-462/miR-731 morphants showed an apparent anterior expansion with an increased expression of etsrp and scl, and an enhanced specification of pu.1-expressing myeloid progenitors. Recent studies showed that etsrp specifically regulates ALPM myelopoiesis besides its previously demonstrated requirement for endothelial lineage [43, 44]. And, scl acts downstream of etsrp to regulate myelopoiesis and vasculogenesis [45,46,47]. The etsrp/scl signaling is critical for myelopoiesis. Overexpression of etsrp is sufficient to induce strong ectopic scl and pu.1 expression in the ALPM, and cause an increased number of l-plastin-expressing leukocytes, while the PLPM erythrogenesis is not significantly affected [43, 44]. We therefore speculate that miR-462-731 acts upstream of etsrp to regulate ALPM myelopoiesis before the myeloid commitment driven by pu.1. In parallel, BMP receptor Alk8 signaling also provides an instructive input for ALPM myelopoiesis and is required for the interplay between pu.1 and gata1, but this function is independent of the role of Alk8 during axis patterning [32]. Also, signaling through alk8 and scl for induction of pu.1-dependent primitive myelopoiesis are considered separate and equally necessary [32, 43]. As expected, miR-462 MO failed to induce pu.1 expression in ALPM in alk8 morphants, supporting the proposition that miR-462-731 acts upstream of alk8 within the BMP signaling. However, alk8 is not required for miR-731 MO to induce ectopic pu.1 expression, suggesting downstream direct regulation between miR-731 and pu.1. Our data further characterized a novel pu.1/miR-462-731 feedback loop which may be responsible for this. There is a cross-inhibitory mechanism between pu.1 and gata1 [19], indicating that the downregulation of gata1 may be determined by the ability of miR-462-731 to regulate pu.1, which leads to deficient erythrogenesis and accentuated myeloid differentiation in miR-462/miR-731 morphants. Together, these results suggest that miR-462-731 functions as a specific modulator of pu.1-dependent ALPM myeloid development and myeloid-erythroid lineage differentiation, acting at a very early stage during zebrafish development.
Notably, a mild ventralization characterized by posterior ICM expansion was detected at 24 hpf. Molecular analysis revealed activation of BMP signaling as early as 70%-epiboly stage with an induced expression of P-Smad1/5 and alk8. High BMP signaling specifies the most ventral mesoderm and results in expansion of blood islands [35, 48]. The Alk8-mediated Bmp2b/7 signaling is known to act maternally and zygotically during dorsoventral patterning of zebrafish embryo [7, 8]. Furthermore, target prediction and functional annotation analysis suggested an involvement of miR-462-731 in BMP/TGF-β signaling by regulating pivotal factors, including Noggins, BMPs, BMPRIs (alk8), Smad1/5/8, and Ids (Supplementary Table S4 and S5). Dorsomorphin, a BMP type I receptor inhibitor, blocks BMP-mediated Smad1/5/8 phosphorylation and may reverse ventralization in chordin morphants [36]. Rescue experiments with both dorsomorphin treatment and alk8 MO co-injection showed that miR-462-731 might act upstream of alk8 within the Bmp2b/7 signaling pathway and function as a novel endogenous BMP antagonist. Therefore, we hypothesize that the ICM expansion in miR-462/miR-731 morphants represents an enhanced hematopoietic specification of mesodermal progenitors that resulted from elevation of BMP signaling.
Besides, our data suggested that a disruption in vascular development may be liable for the striking deficit of blood circulation in miR-462/miR-731 morphants. An impairment of angiogenesis was demonstrated by striking defect in the ISV formation and loss of DLAV integrity. The specification of arteries and veins was also perturbed, as characterized by the irregular patterning of efnb2a, flt4, and flk1 expression. Studies have reported a signaling cascade for arterial fate determination consisting of sequential Hedgehog, Vegf and Notch signaling [49, 50]. In zebrafish, a major role of Notch signaling in blood vessels is to repress venous differentiation within developing arteries [51]. Here, we observed a dramatic reduction of efnb2a and expansion of flt4 into the arterial domain, which is a typical phenotype associated with repression of Notch signaling as previously described [51]. Functional annotation of putative targets suggested that the Notch pathway may be regulated on all layers of the signaling cascade by miR-462-731: single-pass membrane receptor (Notch1), integral membrane ligands (Jagged1/2) and intercellular co-activators/co-repressors (Supplementary Table S5). Using Tg(flk1:GFP/gata1:DsRed), we also detected severely disrupted axial blood circulation in morphants. Specification of HSCs depends on blood flow through the axial circulation [52]. We further identified abnormalities in definitive HSC development. It is reasonable to deduce that miR-462-731 might be indispensable for Notch regulation of arteriovenous programming and may consequently affect blood circulation and downstream HSC production. However, further researches are needed to clarify the detailed regulatory role of miR-462-731 within Notch signaling.
In summary, we for the first time uncover the vital roles of miR-462-731 in regulating embryonic development and hematopoiesis by loss-of-function analysis. We identify miR-462-731 as a novel endogenous BMP antagonist that is required for ventral mesoderm patterning and hematopoietic specification by regulating Alk8-mediated Bmp2b/7 signaling (Fig. 8). Our data reveal that the regulation on etsrp/scl signaling by miR-462-731 combined with the feedback loop between pu.1 and miR-462-731 constitute a regulatory mechanism driving primitive myelopoiesis in the ALPM (Fig. 8). Besides, we suggest a requirement for miR-462-731 in modulating vascular specification and angiogenesis. These findings might facilitate the understanding of miRNA-mediated regulatory mechanism for embryonic hematopoiesis.
Diagrammatic representation of the working model for miR-462-731 to regulate embryonic hematopoiesis. In zebrafish embryos, miR-462-731 regulates dorsoventral mesoderm patterning and hematopoietic specification by regulating Alk8-mediated BMP/Smad signaling. Meanwhile, miR-462-731 provides an instructive input for ALPM myelopoiesis through regulation of etsrp/scl signaling combined with a novel pu.1/miR-462-731 feedback loop. Furthermore, miR-462-731 might regulate arteriovenous programming via Notch signaling. However, further researches are needed to clarify the detailed regulatory role of miR-462-731 in Notch signaling
Materials and methods
Zebrafish maintenance
Zebrafish embryos of wild type and transgenic lines, including Tg(mpo:GFP), Tg(lyz:GFP), Tg(coro1a:GFP), Tg(fli1a:EGFP), and Tg(flk1:GFP/gata1:DsRed), were raised and maintained under standard conditions and staged as described previously [53].
Generation of miR-462-731 mutant zebrafish
The zebrafish miR-462-731 cluster locates on the chromosome 8. The miR-462-731 cluster knockout zebrafish (miR-462-731-KO) was generated using CRISPR/Cas9 technology. The guide RNA target sites for miR-462-731 cluster were designed by the online service of the Web site (http://zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx). Knockout experiment was performed as described previously [54]. For mutant detection, the genomic DNA containing two target sites was amplified, followed by sequencing of the PCR products to identify the genotype. The positive F0 zebrafish were backcrossed with the wildtype zebrafish for generating F1. F1 adult zebrafish with the same genotype (+/−) were intercrossed to generate F2 offspring, which contain wildtype (+ /+), heterozygote (+/−), and homozygote (−/−). The primers for detecting mutants are listed in Supplementary Table S6.
Morpholinos, miRNA duplex, mRNA synthesis, and microinjection
MOs were synthesized by Gene Tools (Philomath, OR, USA) and prepared as 1 mM stock solution. The sequence of MO antisense oligonucleotide miR-462 MO is AGCTGCATTATGGGTTCCGTTACCA, miR-731 MO is CGATCCGGGAGAAAACGTGTCATTC, and both target the mature sequences. The miRNA duplex was purchased from GenePharma (Shanghai, China). Capped pu.1 mRNA was synthesized using mMessage mMachine kit (Ambion, TX, USA). For embryo microinjection, miRNA MOs (1–8 ng), duplex (10 μM) and pu.1 mRNA (100–200 pg) were injected separately or in combination into 1-cell stage zebrafish embryos. Other MO used: 1 ng alk8 MO: ACAACTCCTCAAGTGACTCTCAGCG [43].
RNA sequencing and mRNA profiling
Total RNA was extracted from the MO-injected embryos at 48 hpf using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and cDNA libraries were subjected to Illumina sequencing according to the manufacturer’s protocol (Illumina, CA, USA). The NCBI Sequence Read Archive (SRA) database accession number is SRP116760. DEGs were further subjected to GO and KEGG pathway analysis. Differential expression data were subjected to hierarchical clustering and depicted in a heat map format. TargetScan and MicroCosm algorithms were utilized to predict targets of miR-462 and miR-731. Functional annotation was carried out using KOBAS 3.0.
Drug exposure
Zebrafish embryos injected with miR-462/miR-731 MO were exposed to 4 μM dorsomorphin (Sigma-Aldrich, St. Louis, MO, USA) when developed to shield stage [36], and fixed at 24 hpf for WISH detection.
Quantitative real-time PCR (qRT-PCR)
Total RNA was reverse transcribed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). QRT-PCR was performed using DNA Master SYBR Green I (Roche, Indianapolis, IN, USA) on a Light Cycler 480 System (Roche). The data were analyzed according to the ΔΔCt method using 18s rRNA and U6 snRNA as an internal control for mRNA and miRNA, respectively [55]. All primers were listed in Supplementary Table S6.
Whole-mount in situ hybridization (WISH)
WISH was performed as previously described [56] using the antisense DIG-labelled RNA probes of gata1, pu.1, scl, lmo2, fli1a, etsrp, alas2, hbbe2, hbae1, l-plastin, mpo, c-myb, rag1, flk1, flt4, efnb2a, myoD, pax2a, nkx2.5, and ntl.
O-dianisidine staining
To detect the hemoglobin level, zebrafish embryos at different developmental stages were stained with o-dianisidine (Sigma-Aldrich) staining solution for 15–60 min in the dark as previously described [2].
TUNEL assay and AO staining
To detect DNA fragmentation, TUNEL assay was performed in whole-mount zebrafish embryos. Embryos were manually dechorionated, fixed in 4% paraformaldehyde and dehydrated with methanol overnight at −20 °C. After washing with PBST and digesting with protease K, these embryos were stained using the In situ Cell Death Detection Kit, POD (Roche) according to the manufacturer’s instructions. Whole-mount AO staining in zebrafish embryos was performed as previously reported [55].
Cell culture
The HeLa and ZF4 cell lines (Cell Collection Centre for Freshwater Organisms, Huazhong Agricultural University) were maintained in DMEM and DMEM/F12 1:1 medium (Hyclone, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cells were grown at 37 °C and 28 °C, respectively, in a humidified atmosphere containing 5% CO2.
Plasmid constructs
To generate Pu.1 overexpression construct, zebrafish pu.1 (NM_198062.2) coding sequence was cloned into pCMV-Myc expression vector. Pu.1 responsive elements on the 2 kb region upstream of the miR-462-miR-731 cluster locus were predicted using the JASPAR CORE Vertebrate database [57]. To verify the transcription activity, a series of promoter fragments containing different putative Pu.1 responsive elements were amplified and inserted into the pGL3-Basic vector (Promega). For target validation, the 3’UTR fragments of potential miRNA targets were cloned and inserted into the psiCHECK-2 reporter vector (Promega) to generate the dual-luciferase reporter constructs.
Cell transfection and luciferase reporter assays
For Pu.1 knockdown, ZF4 cells were transfected with Pu.1 siRNA (Sense: 5’-GAGACATGAAGGACAGCATCTdTdT-3’) [58] at a concentration of 120 nM. To overexpress zebrafish Pu.1, HeLa cells were transfected with pCMV-Myc-Pu.1 construct. For promoter activity assay and miRNA target detection, corresponding reporter constructs were transfected into HeLa cells, and the dual-luciferase reporter assay was performed as previously described [31].
Western blotting
Zebrafish embryos and HeLa cells were lysed by pipetting in RIPA lysis buffer (Beyotime Biotechnology, Haimen, Jiangsu, China) with added protease and phosphatase inhibitor cocktail inhibitors (Beyotime Biotechnology). Embryo and cell lysates were analyzed by western blotting as described previously [31]. Primary antibodies used were anti-Cdkn1a (p21), anti-Lmo2 (Homemade polyclonal antibody prepared using prokaryotic expression system, affinity-purified and generated in rabbits), rabbit anti-phospho-Smad1/5 (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-Myc-tag and anti-β-actin (Biodragon Immunotechnologies, Beijing, China).
Microscopy
Live embryos and stained embryos were mounted in 5% methylcellulose and glycerol, respectively. Bright field and fluorescent images were collected using a Leica DFC550 camera driven by the LAS V4.6 software under a Leica M205FA fluorescence stereoscopic microscope. Confocal images were acquired with a Leica TCS SP8 confocal laser microscope equipped with the LAS AF software.
Statistical analysis
The data were represented as mean ± SD (N = 3). The Student’s unpaired 2-tailed t-test was used for all statistical difference evaluation, and P < 0.05 was considered to be statistically significant.
References
Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–44.
Detrich HWr, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA. et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA. 1995;92:10713–7.
Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–56.
Paik EJ, Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol. 2010;54:1127–37.
Warga RM, Nüsslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–38.
Little SC, Mullins MC. Extracellular modulation of BMP activity in patterning the dorsoventral axis. Birth Defects Res C Embryo Today Rev. 2006;78:224–42.
Bauer H, Lele Z, Rauch GJ, Geisler R, Hammerschmidt M. The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development. 2001;128:849–58.
Mintzer KA, Lee MA, Runke G, Trout J, Whitman M, Mullins MC. Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development. 2001;128:859–69.
Snyder A, Fraser ST, Baron MH. Bone morphogenetic proteins in vertebrate hematopoietic development. J Cell Biochem. 2004;93:224–32.
Larsson J, Karlsson S. The role of Smad signaling in hematopoiesis. Oncogene. 2005;24:5676–92.
Sadlon TJ, Lewis ID, D’Andrea RJ. BMP4: its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells. 2004;22:457–74.
Choi K. The hemangioblast: a common progenitor of hematopoietic and endothelial cells. J Hematother Stem Cell Res. 2002;11:91–101.
Hsia N, Zon LI. Transcriptional regulation of hematopoietic stem cell development in zebrafish. Exp Hematol. 2005;33:1007–14.
Berman JN, Kanki JP, Look AT. Zebrafish as a model for myelopoiesis during embryogenesis. Exp Hematol. 2005;33:997–1006.
Crowhurst MO, Layton JE, Lieschke GJ. Developmental biology of zebrafish myeloid cells. Int J Dev Biol. 2002;46:483–92.
Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–51.
Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–45.
Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, et al. Characterization of embryonic globin genes of the zebrafish. Dev Biol. 2003;255:48–61.
Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108.
Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–16.
Lawrie CH. MicroRNAs and haematology: small molecules, big function. Br J Haematol. 2007;137:503–12.
Grabher C, Payne EM, Johnston AB, Bolli N, Lechman E, Dick JE, et al. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2011;25:506–14.
Fan HB, Liu YJ, Wang L, Du TT, Dong M, Gao L, et al. miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood. 2014;124:1320–30.
Lin CY, Lee HC, Fu CY, Ding YY, Chen JS, Lee MH, et al. MiR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat Commun. 2013;4:2829.
Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, Kulshreshtha R. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFbeta-signaling in hypoxic microenvironment. Sci Rep. 2015;5:9650.
Liu L, Zhao Z, Zhou W, Fan X, Zhan Q, Song Y. Enhanced expression of miR-425 promotes esophageal squamous cell carcinoma tumorigenesis by targeting SMAD2. J Genet Genom. 2015;42:601–11.
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68.
Gu Y, Ampofo E, Menger MD, Laschke MW. miR-191 suppresses angiogenesis by activation of NF-κB signaling. FASEB J. 2017;31:3321–33.
Lykken EA, Li QJ. The microRNA miR-191 Supports T cell survival following common γ chain signaling. J Biol Chem. 2016;291:23532–44.
Zhang L, Flygare J, Wong P, Lim B, Lodish HF. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011;25:119–24.
Huang CX, Chen N, Wu XJ, Huang CH, He Y, Tang R, et al. The zebrafish miR-462/miR-731 cluster is induced under hypoxic stress via hypoxia-inducible factor 1alpha and functions in cellular adaptations. FASEB J. 2015;29:4901–13.
Hogan BM, Layton JE, Pyati UJ, Nutt SL, Hayman JW, Varma S, et al. Specification of the primitive myeloid precursor pool requires signaling through Alk8 in zebrafish. Curr Biol. 2006;16:506–11.
Schyth BD, Bela-Ong DB, Jalali SA, Kristensen LB, Einer-Jensen K, Pedersen FS, et al. Two virus-induced microRNAs known only from teleost fishes are orthologues of microRNAs involved in cell cycle control in humans. PLoS One. 2015;10:e0132434.
Leung AY, Mendenhall EM, Kwan TT, Liang R, Eckfeldt C, Chen E, et al. Characterization of expanded intermediate cell mass in zebrafish chordin morphant embryos. Dev Biol. 2005;277:235–54.
Schmid B, Fürthauer M, Connors SA, Trout J, Thisse B, Thisse C, et al. Equivalent genetic roles for bmp7/snailhouse and bmp2b/swirl in dorsoventral pattern formation. Development. 2000;127:957–67.
Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41.
Choi J, Donehower LA. p53 in embryonic development: maintaining a fine balance. Cell Mol Life Sci. 1999;55:38–47.
Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80.
Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311.
Peng WZ, Ma R, Wang F, Yu J, Liu ZB. Role of miR-191/425 cluster in tumorigenesis and diagnosis of gastric cancer. Int J Mol Sci. 2014;15:4031–48.
Zhang BC, Zhou ZJ, Sun L. pol-miR-731, a teleost miRNA upregulated by megalocytivirus, negatively regulates virus-induced type I interferon response, apoptosis, and cell cycle arrest. Sci Rep. 2016;6:28354.
Patterson LJ, Gering M, Eckfeldt CE, Green AR, Verfaillie CM, Ekker SC, et al. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 2007;109:2389–98.
Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–10.
Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10.
Dooley KA, Davidson AJ, Zon LI. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 2005;277:522–36.
Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–11.
Ren X, Gomez GA, Zhang B, Lin S. Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood. 2010;115:5338–46.
Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124:4457–66.
Lawson ND, Vogel AM, Weinstein BM. sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev Cell. 2002;3:127–36.
Swift MR, Weinstein BM. Arterial-venous specification during development. Circ Res. 2009;104:576–88.
Lawson ND, Scheer N, Pham VN, Kim CH, Chitnis AB, Campos-Ortega JA, et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–83.
North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–48.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310.
Zhang D, Wang J, Zhou C, Xiao W. Zebrafish akt2 is essential for survival, growth, bone development, and glucose homeostasis. Mech Dev. 2017;143:42–52.
He Y, Huang CX, Chen N, Wu M, Huang Y, Liu H, et al. The zebrafish miR-125c is induced under hypoxic stress via hypoxia-inducible factor 1α and functions in cellular adaptations and embryogenesis. Oncotarget. 2017;8:73846–59.
Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69.
Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–147.
Wei N, Pang W, Wang Y, Xiong Y, Xu R, Wu W, et al. Knockdown of PU.1 mRNA and AS lncRNA regulates expression of immune-related genes in zebrafish Danio rerio. Dev Comp Immunol. 2014;44:315–9.
Acknowledgements
We are grateful to Prof. Anming Meng, Tsinghua University, for providing the Tg(flk1:GFP/gata1:DsRed) transgenic line. This work was supported by Natural Science Foundation of China (31872542), Top-notch Talent Support Program of College of Fishery, the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and the Fundamental Research Funds for the Central Universities (2016PY030).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by: S. Kumar.
Rights and permissions
About this article
Cite this article
Huang, CX., Huang, Y., Duan, XK. et al. Zebrafish miR-462-731 regulates hematopoietic specification and pu.1-dependent primitive myelopoiesis. Cell Death Differ 26, 1531–1544 (2019). https://doi.org/10.1038/s41418-018-0234-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0234-0
This article is cited by
-
Zebrafish: a convenient tool for myelopoiesis research
Cell Regeneration (2023)