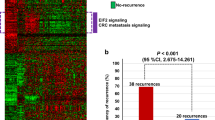
Abstract
Histological grading (HG) is an important prognostic factor of colorectal adenocarcinoma (CRAC): the high-grade CRAC patients have poorer prognosis after tumor resection. Especially, the high-grade stage II CRAC patients are recommended to receive adjuvant chemotherapy. Due to the subjective nature of HG assessment, it is difficult to achieve consistency among pathologists, which brings patients uncertain grading outcomes and inappropriate treatments. We developed a qualitative transcriptional signature based on the within-sample relative expression orderings (REOs) of gene pairs to discriminate high-grade and low-grade CRAC. Using the stage II–III CRAC samples, we detected gene pairs with stable REOs in the high-grade samples and reversal stable REOs in the low-grade samples, and retained the gene pairs whose specific REO patterns were significantly associated with the disease-free survival of patients by univariate Cox regression model. Then, we used a forward-backward searching procedure to extract gene pairs with the highest concordance index as the final grading signature. Finally, 9 gene pairs (9-GPS) were developed to divide CRAC patients into high-grade and low-grade groups. With the signature, there were more differential expression characteristics between reclassified high-grade and low-grade groups. Significant difference of prognosis between the classified two group patients could be seen in four independent datasets. Additionally, genomic analyses showed that the classified high-grade groups were characterized by hypermutation while classified low-grade groups were characterized by frequent copy number alternations. In conclusion, the 9-GPS can provide an objective and robust grading assessment for CRAC patients, which could assist clinical treatment decision.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
05 December 2019
In the original version of this Article, the affiliation details for Department of Systems Biology, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, 150086, China were not assigned to all of the authors. This has now been corrected in both the PDF and HTML versions of the Article.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clinicians. 2018;68:394–424.
Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3:153–73.
Greene FL, Stewart AK, Norton HJ. A new TNM staging strategy for node-positive (stage III) colon cancer: an analysis of 50,042 patients. Ann Surg. 2002;236:416–21. discussion 421
Flejou JF. [WHO Classification of digestive tumors: the fourth edition]. Annales de pathologie. 2011;31(5 Suppl):S27–31.
Compton CC, Fielding LP, Burgart LJ, Conley B, Cooper HS, Hamilton SR, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–94.
Bockelman C, Engelmann BE, Kaprio T, Hansen TF, Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncologica. 2015;54:5–16.
Benson AB 3rd, Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Cancer Netw. 2017;15:370–98.
Berho M, Bejarano PA. Rectal cancer and the pathologist. Minerva chirurgica. 2018;73:534–47.
Chandler I, Houlston RS. Interobserver agreement in grading of colorectal cancers-findings from a nationwide web-based survey of histopathologists. Histopathology. 2008;52:494–9.
Cava C, Bertoli G, Ripamonti M, Mauri G, Zoppis I, Della Rosa PA, et al. Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS ONE. 2014;9:e97681.
Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301.
Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–9.
Cheng J, Guo Y, Gao Q, Li H, Yan H, Li M, et al. Circumvent the uncertainty in the applications of transcriptional signatures to tumor tissues sampled from different tumor sites. Oncotarget. 2017;8:30265–75.
Freidin MB, Bhudia N, Lim E, Nicholson AG, Cookson WO, Moffatt MF. Impact of collection and storage of lung tumor tissue on whole genome expression profiling. J Mol diagnostics. 2012;14:140–8.
Degrelle SA, Hennequet-Antier C, Chiapello H, Piot-Kaminski K, Piumi F, Robin S, et al. Amplification biases: possible differences among deviating gene expressions. BMC Genomics. 2008;9:46.
Qi L, Chen L, Li Y, Qin Y, Pan R, Zhao W, et al. Critical limitations of prognostic signatures based on risk scores summarized from gene expression levels: a case study for resected stage I non-small-cell lung cancer. Brief Bioinf. 2016;17:233–42.
Chen R, Guan Q, Cheng J, He J, Liu H, Cai H, et al. Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples. Oncotarget. 2017;8:6652–62.
Song K, Guo Y, Wang X, Cai H, Zheng W, Li N, et al. Transcriptional signatures for coupled predictions of stage II and III colorectal cancer metastasis and fluorouracil-based adjuvant chemotherapy benefit. FASEB J. 2019;33:151–62.
Guan Q, Yan H, Chen Y, Zheng B, Cai H, He J, et al. Quantitative or qualitative transcriptional diagnostic signatures? A case study for colorectal cancer. BMC Genomics. 2018;19:99.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic acids Res. 2013;41(Database issue):D991–5.
International Cancer Genome C, Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, et al. International network of cancer genome projects. Nature. 2010;464:993–8.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41.
Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8.
Harrell FE Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87.
Lusted LB. Signal detectability and medical decision-making. Science. 1971;171:1217–9.
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–16 e11.
Bland JM, Altman DG. The logrank test. BMJ. 2004;328:1073.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids Res. 2012;40(Database issue):D109–14.
Blenkinsopp WK, Stewart-Brown S, Blesovsky L, Kearney G, Fielding LP. Histopathology reporting in large bowel cancer. J Clin Pathol. 1981;34:509–13.
Kakizaki F, Aoki K, Miyoshi H, Carrasco N, Aoki M, Taketo MM. CDX transcription factors positively regulate expression of solute carrier family 5, member 8 in the colonic epithelium. Gastroenterology. 2010;138:627–35.
Qualtrough D, Hinoi T, Fearon E, Paraskeva C. Expression of CDX2 in normal and neoplastic human colon tissue and during differentiation of an in vitro model system. Gut. 2002;51:184–90.
Blaker H. Grading of tumors in the tubular digestive tract: Esophagus, stomach, colon and rectum. Der Pathol. 2016;37:293–8.
Lee M, Rhee I. Cytokine signaling in tumor progression. Immune Netw. 2017;17:214–27.
Tzanakakis G, Kavasi RM, Voudouri K, Berdiaki A, Spyridaki I, Tsatsakis A, et al. Role of the extracellular matrix in cancer-associated epithelial to mesenchymal transition phenomenon. Developmental Dyn. 2018;247:368–81.
Rizeq B, Zakaria Z, Ouhtit A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 2018;109:33–42.
PN S, Darvin P, Yoo YB, Joung YH, Kang DY, Kim DN, et al. The combination of methylsulfonylmethane and tamoxifen inhibits the Jak2/STAT5b pathway and synergistically inhibits tumor growth and metastasis in ER-positive breast cancer xenografts. BMC cancer. 2015;15:474.
Jasmine F, Rahaman R, Dodsworth C, Roy S, Paul R, Raza M, et al. A genome-wide study of cytogenetic changes in colorectal cancer using SNP microarrays: opportunities for future personalized treatment. PLoS ONE. 2012;7:e31968.
Xu WQ, Jiang XC, Zheng L, Yu YY, Tang JM. Expression of TGF-beta1, TbetaRII and Smad4 in colorectal carcinoma. Exp Mol Pathol. 2007;82:284–91.
Brozek W, Manhardt T, Kallay E, Peterlik M, Cross HS. Relative expression of vitamin D hydroxylases, CYP27B1 and CYP24A1, and of Cyclooxygenase-2 and heterogeneity of human colorectal cancer in relation to age, gender, tumor location, and malignancy: results from factor and cluster analysis. Cancers. 2012;4:763–76.
Olender J, Nowakowska-Zajdel E, Kruszniewska-Rajs C, Orchel J, Mazurek U, Wierzgon A, et al. Epigenetic silencing of endothelin-3 in colorectal cancer. Int J Immunopathol Pharmacol. 2016;29:333–40.
Massague J. TGFbeta in. Cancer Cell. 2008;134:215–30.
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. New Engl J Med. 2009;361:2449–60.
Yu Y, Liu D, Liu Z, Li S, Ge Y, Sun W, et al. The inhibitory effects of COL1A2 on colorectal cancer cell proliferation, migration, and invasion. J Cancer. 2018;9:2953–62.
Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol. 2008;384:766–79.
Schneider F, Kemmner W, Haensch W, Franke G, Gretschel S, Karsten U, et al. Overexpression of sialyltransferase CMP-sialic acid:Galbeta1,3GalNAc-R alpha6-Sialyltransferase is related to poor patient survival in human colorectal carcinomas. Cancer Res. 2001;61:4605–11.
Bader AG, Byrom M, Johnson CD, Brown D. MIR-143 regulated genes and pathways as targets for therapeutic intervention. U.S. Patent Application. 2008.
Lee YS, Kim SY, Song SJ, Hong HK, Lee Y, Oh BY, et al. Crosstalk between CCL7 and CCR3 promotes metastasis of colon cancer cells via ERK-JNK signaling pathways. Oncotarget. 2016;7:36842–53.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers: 61601151, 61701143, 61673143, 81872396 and 81572935], the Natural Science Foundation of Heilongjiang Province [grant number: C2016037] and the Joint Scientific and Technology Innovation Fund of Fujian Province [grant number: 2016Y9044].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, H., Song, K., Fu, Y. et al. A qualitative transcriptional signature for determining the grade of colorectal adenocarcinoma. Cancer Gene Ther 27, 680–690 (2020). https://doi.org/10.1038/s41417-019-0139-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-019-0139-1
This article is cited by
-
Development of machine learning-based predictors for early diagnosis of hepatocellular carcinoma
Scientific Reports (2024)
-
Identifying individualized prognostic signature and unraveling the molecular mechanism of recurrence in early-onset colorectal cancer
European Journal of Medical Research (2023)
-
StemSC: a cross-dataset human stemness index for single-cell samples
Stem Cell Research & Therapy (2022)
-
A novel qualitative signature based on lncRNA pairs for prognosis prediction in hepatocellular carcinoma
Cancer Cell International (2022)