Abstract
Background
Wisteria floribunda agglutinin (WFA)+ Mac-2-binding protein (M2BPGi) is a novel serum marker for liver fibrosis. Although an elevated serum level of M2BPGi can predict development of hepatocellular carcinoma (HCC), the effect of M2BPGi on HCC remains unclear. There are no reports about the association of M2BPGi with HCC aggressiveness. We aimed to clarify the significance of M2BPGi in HCC.
Methods
The protein expression of M2BPGi and galectin-3, a ligand of M2BP, and the mRNA expression of M2BP were evaluated in surgically resected human HCC samples. M2BPGi-regulating signals in HCC cells were investigated using transcriptome analysis. The effects of M2BPGi on HCC properties and galectin-3/mTOR signaling were evaluated.
Results
M2BPGi and galectin-3 proteins co-localised in HCC cells, while M2BP mRNA was detected in cirrhotic liver stromal cells. mTOR signaling was upregulated in M2BPGi-treated HCC cells. Moreover, M2BPGi treatment induced tumour-promoting effects on HCC in vitro by activated mTOR signaling. In addition, M2BPGi bound to galectin-3 to induce membranous galectin-3 expression in HCC cells. In vivo, M2BPGi enhanced the growth of xenografted HCC.
Conclusions
M2BPGi is produced in stromal cells of the cirrhotic liver. Furthermore, M2BPGi enhances the progression of HCC through the galectin-3/mTOR pathway.
Similar content being viewed by others
Background
Liver cancer is the sixth and ninth most common type of cancer in men and women, respectively, and the fourth most common cause of cancer-related death worldwide. The majority of liver cancers involve the histological subtype hepatocellular carcinoma (HCC), a primary malignancy of the liver.1 Several liver factors, particularly fibrosis, contribute to the risk of hepatocarcinogenesis. Approximately 70–90% of patients with HCC have a history of chronic liver disease and cirrhosis,2 and patients with liver cirrhosis (Child–Pugh score B or C) who develop HCC have a significantly worse prognosis than those without cirrhosis.3 Although liver resection is the most effective treatment for patients with early HCC who have a non-cirrhotic or mildly cirrhotic liver,4 few treatment options are available for those with severe liver fibrosis.5 Therefore, a deeper understanding of HCC in fibrotic liver is essential for its clinical management.
According to Kuno et al., Wisteria floribunda agglutinin (WFA) lectin can bind to terminal N-acetylgalactosamine motifs on carbohydrate structures, as well as with Mac-2-binding protein (M2BP, also known as galectin-3-binding protein) glycoforms during liver fibrosis.6 Accordingly, WFA-binding M2BP has been termed WFA+ M2BP or Mac-2-binding protein glycan isomer (M2BPGi).6 Clinical studies have identified M2BPGi as a novel serum marker of liver fibrosis associated with various aetiologies, including hepatitis B and C virus infections, non-alcoholic fatty liver disease and primary biliary cirrhosis,7,8,9,10,11 and the serum M2BPGi level may predict HCC development and prognosis in patients with hepatitis B and C virus infections.12,13,14,15 Bekki et al. reported that M2BPGi is secreted by hepatic stellate cells (HSCs) in the cirrhotic liver and subsequently activates Kupffer cells to promote hepatic fibrosis.16 Although a few studies addressed the source of M2BPGi in the cirrhotic liver, no basic studies have explained the potential association of M2BPGi with HCC aggressiveness.
mTOR signaling is upregulated in 40–50% of HCCs,17 and this pathway has been studied for the development of molecular targeted therapies for several cancers, including HCC.18 Interestingly, mTOR signaling is regulated by galectin-3,17,18 a well-known binding partner of M2BP protein. Although the expression of galectin-3 was reported to be higher in HCC than non-cancerous tissues, and to associate with a poor prognosis,19,20,21 the relationship between mTOR signaling, galectin-3 and M2BPGi in HCC remains unknown.
The investigations in the present study were conducted to clarify the source of M2BPGi in the liver, and the fundamental effects of M2BPGi against HCC cells. Specifically, the localisation of glycosylated M2BP as M2BPGi, galectin-3 and M2BP mRNA was evaluated in HCC tissues resected from fibrotic liver, and the tumour-promoting effects of M2BPGi were analysed in vitro and in vivo. Furthermore, Cap Analysis of Gene Expression (CAGE) was used to identify the relationship between M2BPGi and the novel downstream factors of galectin-3 and mTOR signaling in HCC cells.
Methods
Multicolour immunofluorescence staining
Five surgically resected HCC specimens were fixed with 1% formaldehyde, embedded in paraffin, cut into 4-µm-thick sections and mounted on glass slides. Deparaffinisation, antigen retrieval, endogenous peroxidase blocking and blocking protocols were described previously.22 The sections were incubated with primary antibodies, including rat anti-galectin-3 antibody (Cedarlane), goat anti-M2BP antibody (R&D Systems) and biotinylated WFA lectin (Vector Laboratories), and the appropriate Alexa Fluor-conjugated secondary antibodies and fluorescent streptavidin conjugates (1:1000, Invitrogen) for 60 min per incubation. The cell nuclei were labelled with Spectral DAPI (PerkinElmer). The labelled cells were visualised using a BZ-X710 fluorescent microscope (Keyence).
RNA in situ hybridisation assay
Three paraffin-embedded tissue sections on slides were subjected to RNA in situ hybridisation to detect the localisation and expression of M2BP RNA, using a M2BP-specific probe and RNAscopeR2.5 HD Detection kit (BROWN; ACD Bio-Techne). The technique was performed manually according to the manufacturer’s protocol. The resulting images were evaluated using a light microscope (Carl Zeiss Axio Scope.A1 microscope).
Cell culture
To validate the importance and function of M2BPGi in several differentiated cell lines of HCC, we selected the PLC/PRF/5 cell line to represent poorly differentiated HCC,23 whereas other cell lines (Huh7 and HepG2) represented well-differentiated HCC.24 All cell lines were obtained from the JCRB Cell Bank (Osaka, Japan). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing L-glutamine, Phenol Red and Sodium Pyruvate (Fujifilm Wako Pure Chemical Corporation) and supplemented with 10% foetal bovine serum (FBS) and 1% penicillin–streptomycin (ThermoFisher Scientific), and were incubated at 37 °C in a humidified incubator at 5% CO2.
Proliferation assay
Cell-proliferation ability was analysed using Cell counting kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan). HCC cells were seeded at a density of 3000 cells/well in 96-well culture plates. After 24 h, the medium was changed with and without 1 or 3 µg/ml M2BPGi (Sysmex Co.), sugar chain cut-off—M2BPGi (SC-M2BPGi) and 10 nM mTOR inhibitor (rapamycin, EMD Millipore). Cell proliferation was evaluated at initial seeding (0 h) and at 24, 48 and 72 h of treatment. The absorbance of each well was measured using a spectrophotometer (Bio-Rad, Hercules, CA, USA) at 450 nm with the reference wavelength set at 650 nm. All experiments were performed in replicates of six.
Invasion assay
The cell-invasion assay was performed using 24-well Corning® BioCoat™ Matrigel Invasion Chambers (Corning, NY, USA). HCC cell lines in FBS-free medium were seeded into the Matrigel inserts at a density of 1 × 105 per chamber. The lower chamber contained 1 or 3 µg/ml M2BPGi in medium containing 10% FBS. After incubation for 48 h, the cells were fixed and stained with Diff-Quik (Sysmex Corporation, Kobe, Japan). After staining, cells that migrated through the pores to the lower surface of the membrane were counted under the microscope. In total, five randomly selected fields were evaluated.
Wound-healing assay
Cell-migration ability was evaluated in M2BPGi non-treated and treated HCC cells (PLC/PRF/5) using wound-healing assay. The cells were seeded in the six-well plates and cultured until reaching confluence. A uniform straight wound was generated using pipette tip. Each well was washed with PBS to remove detached cells, and wells were cultured in medium with and without M2BPGi (3 µg/ml) at 37 °C for 48 h. The cell-migration rate was calculated using replicates of five.
In vivo experiment
Six female NOD-SCID mice (7 weeks old, body weight: 19–20 g) were purchased from CLEA Japan, Inc. Mice have free access to water and food, housed in specific pathogen-free cages and bedding in a 12-h light/dark regimen with controlled room temperature. PLC/PRF/5 cells were suspended in 10% FBS DMEM at a density of 5 × 106/200 µl and injected subcutaneously into both flanks of each mouse. Each group contained five xenografts. After 7 days, the mice were randomly divided into control (PBS) and M2BPGi groups and implanted subcutaneously with mini-osmotic pumps (Alzet®, model 2002) charged with PBS or M2BPGi (303 µg/ml), respectively. M2BPGi was secreted subcutaneously with 3.6 µg/ml a day for 14 days. During the procedures of injection and pump implantation, mice were anaesthetised with isoflurane (Pfizer Japan Inc., Tokyo). The mice were observed until complete recovery from anaesthesia. The tumour sizes and body weights were measured every 2 days after pump implantation, and the tumour volumes were calculated using the following formula: V = (W2 × L)/2. To collect the xenografted tumours, mice were deeply anaesthetised with isoflurane and euthanised by cervical dislocation on the 14th day of M2BPGi administration. The resected tumours were fixed with formalin-neutral buffer 20 (Kenei Pharmaceutical, Japan) for pathological examination. All mouse experiments were conducted in compliance with the guidelines of the Institute for Laboratory Animal Research at Gunma University, Maebashi, Japan (Approval number: 18-024).
Immunohistochemistry
Immunohistochemical staining was performed using antibodies specific for Ki-67 (Dako, Agilent Technologies) and phospho-S6 (anti-rabbit, Cell Signaling Technology) according to previously described methods.22 Ki-67-positive cancer cells from samples treated with PBS and M2BPGi were counted directly on 10 representative images at the same magnification (×400).
siRNA transfection
HCC cells were suspended in Opti-MEM I-reduced Serum Media (ThermoFisher Scientific) at a density of 1.0 × 106/100 µl and mixed with control siRNA (Dharmacon) or the following galectin-3 siRNAs at a concentration of 200 µM: #1, sense: 5′-GGAGAGUCAUUGUUUGCAA-3′, antisense: 5′-UUGCAAACAAUGACUCUCC-3′; siRNA #2, sense: 5′-GGGAAUGAUGUUGCCUUCCACUUUA-3′, antisense: 5′-UAAAGUGGAAGGCAACAUCAUUCCC-3′. All siRNA transfections were performed via electroporation using a CUY21 EDIT II electroporator (BEX) at poring and transfer pulses of 150 and 10 V, respectively.
Protein extraction and Western blotting analysis
HCC cell lines were treated with M2BPGi at concentrations varying between 0 µg/ml and 3 µg/ml for 6 h. After 24 h of galectin-3 siRNA transfection, cells were treated with 1 µg/ml of M2BPGi for 24 h, followed by protein extraction. Protein extraction was performed using RIPA buffer (Wako) containing phosphatase inhibitor (p2850, Sigma-Aldrich) according to the manufacturer’s instructions.
Membrane proteins were extracted using a Mem-PER Plus Membrane protein extraction kit (ThermoFisher Scientific) according to the manufacturer’s protocol. An approximately 2-mm section of xenograft tissue was homogenised in RIPA buffer (Wako) containing phosphatase inhibitor (p285, Sigma-Aldrich) according to the manufacturer’s protocol.
Proteins were separated using SDS-PAGE with 10% Bis–Tris gels and transferred to nitrocellulose membranes (#12369, Cell Signaling). Membranes were blocked with 5% skimmed milk or 5% BSA and incubated overnight at 4 °C with the primary antibodies. Primary antibodies used in the present study were anti-galectin-3 (Cedarlane), phospho-mTOR, mTOR, phospho-p70–S6K, p70–S6K, phospho-S6 (Cell Signaling Technology) and beta-actin (Sigma). Next, membranes were treated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Protein bands on the membrane were detected using ECL™ Prime Western Blotting Detection Reagent and an ImageQuant™ LAS 4000 imager (GE Healthcare, Buckinghamshire, UK).
Co-immunoprecipitation
PLC/PRF/5 cells were incubated with or without 3 µg/ml M2BPGi for 12 h and subsequently collected and lysed with 1% Triton-X. For co-immunoprecipitation, a lysate volume containing 300 µg of protein was incubated with 3 µl of human galectin-3 antibody (R&D Systems) for 2 h at 4 °C with constant rotation. Subsequently, 20 µl of protein G beads were added, and the mixture was incubated for another 1 h with constant rotation. After 5 washes, the precipitated proteins were resuspended in 20 µl of SDS sample buffer and boiled at 95 °C for 5 min. Protein expression was evaluated by western blotting.
Cap analysis of gene expression
PLC/PRF/5 cells were seeded in 10-cm dishes and grown to 70–80% confluency at 37 °C in a humidified incubator with 5% CO2. The medium was supplemented or not with 3 µg/ml M2BPGi. After 6 h, the cells were collected and washed with PBS, snap-frozen in nitrogen and stored at −80 °C. The CAGE library preparation, sequencing, mapping and gene expression and motif discovery analysis were performed by DNAFORM. Briefly, RNA quality was assessed using a Bioanalyzer (Agilent) to ensure that the RNA-integrity number (RIN) exceeded 7.0, and the A260/280 and A260/230 ratios exceeded 1.7. First-strand cDNAs were transcribed to the 5′ ends of capped RNAs and attached to CAGE ‘bar code' tags. The sequenced CAGE tags were then mapped to mouse mm9 genomes using BWA software (v0.5.9) after discarding ribosomal or non-A/C/G/T base-containing RNAs. Finally, the CAGE-tag 5′ coordinates were input for CAGEr clustering25 using the Paraclu algorithm26 with the default parameters.
Statistical analysis
The experimental data of the proliferation, invasion and xenografted tumour-volume assays were analysed using replicates of 5–6. Data for continuous variables are expressed as means ± standard deviations (SDs). Differences among groups were evaluated using the t test and Tukey’s test. In vivo data were analysed using repeated-measures analysis of variance. The results with P values < 0.05 were considered statistically significant. All statistical analyses were conducted using the JMP 13.0.0 software package (SAS Institute Inc.).
Results
Co-localisation of M2BPGi and galectin-3
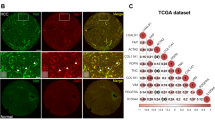
In a previous study, M2BPGi-positive cells were investigated in fibrotic liver samples using immunohistochemical analysis.16 In our study, we evaluated M2BPGi expression in HCC tissue samples using immunohistochemistry. The results revealed that M2BPGi and galectin-3 were co-localised in HCC cells (Fig. 1a). In addition, adjacent non-tumour areas were analysed. M2BPGi co-localised with a Kupffer cell marker (CD68) (Supplementary Fig. 1) and galectin-3 (Fig. 1b) according to immunofluorescence staining.
Representative multi-immunofluorescence images of cells labelled to indicate M2BP (red, upper left panel), WFA (green, upper middle panel), M2BP with WFA as M2BPGi (merged, upper right panel), galectin-3 (pink, lower right panel) or M2BPGi and galectin-3 (merged, lower middle panel) and stained with haematoxylin and eosin (lower right panel) in the tumour (a) and adjacent non-tumour (b) areas of resected hepatocellular carcinoma tissue (×400 magnification).
Expression of M2BP mRNA
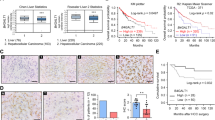
In addition, to investigate the source of M2BP protein, we examined M2BP mRNA expression using an in situ hybridisation assay. M2BP mRNA expression was detected in the stroma of the cirrhotic liver rather than in hepatocytes (Fig. 2a). In addition, we analysed M2BP mRNA expression in HCC tissue samples. M2BP mRNA expression was detected in the stroma of HCC tissues, but not in HCC cells (Fig. 2b). According to the result, we examined M2BP protein expression in HCC cell lines, including PLC/PRF/5, HepG2 and Huh7 cells. M2BP was expressed in PLC/PRF/5 and HepG2 cells, but not in Huh7 cells (Supplementary Fig. 2).
a The expression of M2BP mRNA was assessed using RNA in situ hybridisation. M2BP mRNA was detected (arrows) in stromal cells of the cirrhotic liver (b, c) but not in hepatocytes (e, f) at ×400 magnification. Haematoxylin–eosin staining (a, d) at ×100 magnification. b M2BP mRNA was detected (arrows) in stromal cells of the HCC but not in HCC cells (b, d) at ×400 magnification. Haematoxylin–eosin staining (a, c) at ×100 and ×200 magnification.
M2BPGi treatment and HCC progression
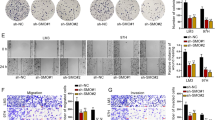
M2BPGi significantly enhanced the proliferation rates of PLC/PRF/5 human HCC cell lines when compared with non-treated cells (Fig. 3a). Other HCC cell lines (HepG2 and Huh7) showed the same result (Supplementary Fig. 3a). On the other hand, sugar chain cut-off M2BPGi (SC-M2BPGi) had no proliferation effect on PLC/PRF/5 cell line (Supplementary Fig. 4). M2BPGi also significantly enhanced the invasiveness and migration of the PLC/PRF/5 cell line when compared with non-treated cells (Fig. 3b) (Supplementary Fig. 3c). HepG2 and Huh7 cell lines showed similar results with PLC/PRF/5 cell line (Supplementary Fig. 3b). In vivo, we confirmed that treatment with M2BPGi enhanced tumour growth significantly in an HCC-bearing mouse model, compared with controls (Fig. 3c). M2BPGi supported the proliferation of HCC cell lines in vitro; therefore, we analysed the expression of the proliferation marker Ki-67 in the xenografted tumours. The Ki-67 labelling index was higher in M2BPGi-treated than in PBS-treated tumours (Fig. 3d).
HCC cell lines (PLC/PRF/5) treated with M2BPGi were subjected to a a proliferation assay (n = 6, *p < 0.05) and b invasion assay (n = 5, *p < 0.05). c M2BPGi significantly enhanced the growth of xenografted tumours when compared with controls (PBS) (n = 5, *p = 0.0023). d The Ki-67 labelling index was significantly higher in M2BPGi-treated xenograft tumours than in controls (n = 10, *p < 0.01) (×400 magnification).
Membranous galectin-3 and tumour-promoting effect of M2BPGi
Next, we focused on the relationship between M2BPGi as a tumour promoter and galectin-3 as a representative M2BP-binding protein. Notably, M2BPGi treatment did not enhance the proliferation of galectin-3–siRNA-knockdown HCC cells relative to controls (Fig. 4a) (Supplementary Fig. 5). Moreover, M2BPGi did not enhance the invasive ability of galectin-3-downregulated cells compared with the findings in the controls (Fig. 4b). To further evaluate the interaction of M2BPGi with galectin-3 in HCC cells, we subjected the PLC/PRF/5 HCC cell line to immunoprecipitation, and observed stronger M2BP expression (85–97 kDa) in M2BPGi-treated cells than in non-treated cells. Glycosylation at N-linked sites of M2BP generates molecular size of 85–97 kDa,27 which represents M2BPGi. Our result indicates that extrinsic M2BPGi could bind to cellular galectin-3 (Fig. 4c). Furthermore, the membranous expression of galectin-3 increased in response to M2BPGi treatment (Fig. 4d).
a Compared with control, M2BPGi did not enhance the proliferation of HCC cell lines (PLC/PRF/5) transfected with Gal-3 siRNA (n = 6, *p < 0.01). Western blotting validated the suppression of Gal-3 by specific siRNAs. b M2BPGi did not enhance the invasion of HCC cell lines (PLC/PRF/5) transfected with Gal-3 siRNA compared with controls (n = 5, *p < 0.01). c M2BP immunoprecipitation using a Gal-3-specific antibody (Gal-3 IP)-detected M2BP at 75-kDa band in whole-cell lysate. M2BPGi-treated Gal-3 IP samples, but not controls, appeared as a broad M2BP band at 85–97 kDa. d Membranous Gal-3 protein expression in PLC/PRF/5 cells treated with M2BPGi. E cadherin was used as a membrane protein- loading control.
M2BPGi and mTOR signaling in HCC cells
To clarify the mechanism by which M2BPGi promotes HCC, we subjected PLC/PRF/5 cells to a CAGE analysis. A gene ontology analysis suggested that mTOR signaling was upregulated in M2BPGi-treated cells relative to non-treated cells (Fig. 5a). Therefore, we evaluated the induction of mTOR, p70–S6 kinase and S6 as representative downstream components of the mTOR- signaling pathway in M2BPGi-treated cells. We further validated the dose-dependent induction of mTOR, p70–S6 kinase and S6 phosphorylation in M2BPGi-treated cells by western blotting (Fig. 5b).
a Enhanced mTOR signaling in M2BPGi-treated PLC/PRF/5 cells by GO analyses. b M2BPGi induced the phosphorylation of mTOR, p70–S6K and S6 in PLC/PRF/5 cells. c Phospho-S6 was enhanced in the lysates of M2BPGi-treated xenograft tumour tissues. Representative images of phospho-S6 staining (×400 magnification). d Gal-3 downregulation reduced the expression of mTOR -signaling pathway. e The proliferative effects of M2BPGi on HCC cells were inhibited by treatment with rapamycin (mTOR inhibitor) (n = 6, *p < 0.01). f Schema of the mTOR-meditated functions of M2BPGi.
Consistent with our in vitro data, we also observed enhanced levels of phospho-S6, an indicator of activated mTOR signaling, in the M2BPGi-treated xenograft tumours relative to the controls (Fig. 5c). We next evaluated whether galectin-3 was required for the activation of mTOR signaling in HCC cells treated with M2BPGi. We observed downregulated mTOR signaling in the galectin-3–siRNA-knockdown cells relative to the levels in control cells, regardless of the M2BPGi treatment status (Fig. 5d). Finally, although M2BPGi treatment enhanced the proliferation of HCC cells (Fig. 3a), the tumour-promoting effects of M2BPGi were cancelled by treatment with the representative mTOR inhibitor rapamycin (Fig. 5e).
Discussion
In this study, we focused on the liver fibrosis marker, M2BPGi, a glycosylated galectin-3-binding protein derived from the stromal cells of cirrhotic liver. We further showed that M2BPGi could bind to and induce the membrane expression of galectin-3. Moreover, M2BPGi promoted HCC malignancy in vitro via galectin-3, which consequently activated mTOR signaling. This was confirmed by the observation that galectin-3 knockdown significantly inhibited the effect of M2BPGi on HCC cells. Our in vivo findings were consistent with the in vitro data and further demonstrated the importance of M2BPGi-induced mTOR signaling on HCC growth.
To date, to the best of our knowledge, no study has clarified the source of M2BPGi in fibrotic liver tissues. Accordingly, this report is the first to confirm a discrepancy between the localisation of M2BPGi protein and M2BP mRNA in clinical samples. In a previous study, an anti-M2BP antibody and WFA lectin were used to detect glycosylated M2BP protein (i.e. M2BPGi) via staining only in human tissue samples.16 The protein localisation data suggested the importance of M2BPGi in the fibrotic liver.16 In addition, they described the origin of M2BPGi in the fibrotic liver using HSCs, Kupffer cells, biliary epithelial cells and endothelial cells, which were isolated from human fibrotic liver tissues. The results illustrated that HSCs dominantly secreted M2BPGi, thereby activating Kupffer cells in vitro to facilitate liver fibrosis.16 Moreover, M2BPGi has been used as a clinical serum biomarker of liver fibrosis,28 a poor prognosticator in patients with HCC.3,29 Our immunohistochemical data revealed that M2BPGi expression was observed in HCC cells within the tumour areas, and within Kupffer cells rather than hepatocytes in the adjacent non-tumour areas. Therefore, to clarify the source of M2BPGi in the liver tissues, we identified the stromal cells of the cirrhotic liver, rather than the hepatocytes, as the M2BP-producing cells using RNA in situ hybridisation. Taken together, these data suggest that M2BPGi is secreted from stromal cells and may be taken up by HCC cells in the fibrotic liver.
Previously, M2BP protein was reported to affect cancer aggressiveness. In human cancers, M2BP was shown to bind to galectin-3,30,31 which was related to the activation of mTOR signaling in cancer cells.17 However, glycosylation of M2BP was not reported in previous studies. The suppression of galectin-3 downregulated the activation of mTOR and S6K in colon cancer.18 In this study, we used CAGE analysis to identify mTOR signaling as a downstream effector of M2BPGi in HCC, and validated that M2BPGi could bind to galectin-3 and activate mTOR signaling in HCC cells through both in vitro and in vivo experiments. Moreover, mTOR inhibitor treatment and galectin-3 suppression blocked the effects of M2BPGi on HCC aggressiveness. Therefore, our results suggested that M2BPGi may enhance HCC aggressiveness by activating the galectin-3/mTOR axis.
In this study, the activation of mTOR signaling by M2BPGi/galectin-3 was reported; however, the fundamental mechanism by which galectin-3 activates mTOR was not evaluated. In another study, galectin-3 was reported to activate MAPK signaling, which plays a key role in the progression of cancers, including HCC.32,33 Conversely, MAPK signaling has been reported to be upstream of mTOR in lung cancer cells.34 Taken together, these reports suggest that M2BPGi-induced galectin-3 enhances mTOR signaling through MAPK signaling. Although mTOR inhibitors have been developed and used in clinical trials for HCC treatment in the past decade, the tumour-suppressive effects of these agents in patients with HCC remain controversial.35,36 One randomised, multicenter, multinational Phase 2 trial reported that the addition of an mTOR inhibitor to sorafenib did not improve the clinical efficacy compared with sorafenib alone in patients with advanced HCC.36 Moreover, several researchers reported activated mTOR signaling in HCC,37,38,39 and clinical studies identified mTOR activation as a potential indicator of a poor prognosis in patients with HCC.40 In our study, we demonstrated that M2BPGi could enhance the aggressiveness of HCC by activating mTOR signaling in the presence of galectin-3. Our data suggest that the tumour expression of galectin-3 may be a predictive factor of treatment outcomes, and could be used to select patients who would benefit from mTOR inhibitor therapy. Our data further suggest that M2BPGi may regulate the magnitude of mTOR activation in HCC; in other words, the serum level of M2BPGi may reflect the level of mTOR activation in HCC cells. Potentially, M2BPGi may be a useful non-invasive marker for the identification of patients with HCC with mTOR activation. In the future, a therapeutic strategy based on mTOR inhibitors might be useful for the treatment of mTOR-activated HCC associated with a high level of M2BPGi.
In conclusion, M2BPGi is produced by stromal cells in the cirrhotic liver, where it enhances the progression of HCC by activating mTOR signaling in the presence of galectin-3. M2BPGi may serve as a link between HCC and stromal cells, and may thus explain the fundamental mechanism underlying HCC progression. Accordingly, M2BPGi may be a promising therapeutic target in patients with liver cirrhosis who develop HCC.
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A. & Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Blachier, M., Leleu, H., Peck-Radosavljevic, M., Valla, D. C. & Roudot-Thoraval, F. The burden of liver disease in Europe: a review of available epidemiological data. J. Hepatol. 58, 593–608 (2013).
Greten, T. F., Papendorf, F., Bleck, J. S., Kirchhoff, T., Wohlberedt, T., Kubicka, S. et al. Survival rate in patients with hepatocellular carcinoma: a retrospective analysis of 389 patients. Br. J. Cancer 92, 1862–1868 (2005).
Mohkam, K., Dumont, P. N., Manichon, A. F., Jouvet, J. C., Boussel, L., Merle, P. et al. No-touch multibipolar radiofrequency ablation vs. surgical resection for solitary hepatocellular carcinoma ranging from 2 to 5 cm. J. Hepatol. 68, 1172–1180 (2018).
Kamarajah, S. K. Fibrosis score impacts survival following resection for hepatocellular carcinoma (HCC): a surveillance, end results and epidemiology (SEER) database analysis. Asian J. Surg. 41, 551–561 (2018).
Kuno, A., Ikehara, Y., Tanaka, Y., Ito, K., Matsuda, A., Sekiya, S. et al. A serum “sweet-doughnut” protein facilitates fibrosis evaluation and therapy assessment in patients with viral hepatitis. Sci. Rep. 3, 1065 (2013).
Hasegawa, K., Takata, R., Nishikawa, H., Enomoto, H., Ishii, A., Iwata, Y. et al. Impact of Wisteria floribunda agglutinin-positive Mac-2-binding protein in patients with hepatitis C virus-related compensated liver cirrhosis. Int. J. Mol. Sci. 17, 1500 (2016).
Nishikawa, H., Enomoto, H., Iwata, Y., Kishino, K., Shimono, Y., Hasegawa, K. et al. Clinical significance of serum Wisteria floribunda agglutinin positive Mac-2-binding protein level in non-alcoholic steatohepatitis. Hepatol. Res. 46, 1194–1202 (2016).
Nishikawa, H., Hasegawa, K., Ishii, A., Takata, R., Enomoto, H., Yoh, K. et al. A proposed predictive model for advanced fibrosis in patients with chronic hepatitis B and its validation. Medicine (Baltimore) 95, e4679 (2016).
Toshima, T., Shirabe, K., Ikegami, T., Yoshizumi, T., Kuno, A., Togayachi, A. et al. A novel serum marker, glycosylated Wisteria floribunda agglutinin-positive Mac-2 binding protein (WFA(+)-M2BP), for assessing liver fibrosis. J. Gastroenterol. 50, 76–84 (2015).
Umemura, T., Joshita, S., Sekiguchi, T., Usami, Y., Shibata, S., Kimura, T. et al. Serum Wisteria floribunda agglutinin-positive Mac-2-binding protein level predicts Liver fibrosis and prognosis in primary biliary cirrhosis. Am. J. Gastroenterol. 110, 857–864 (2015).
Kim, S. U., Heo, J. Y., Kim, B. K., Park, J. Y., Kim, D. Y., Han, K. H. et al. Wisteria floribunda agglutinin-positive human Mac-2 binding protein predicts the risk of HBV-related liver cancer development. Liver Int. 37, 879–887 (2017).
Sato, S., Genda, T., Ichida, T., Amano, N., Sato, S., Murata, A. et al. Prediction of hepatocellular carcinoma development after hepatitis C virus eradication using serum wisteria floribunda agglutinin-positive Mac-2-binding protein. Int. J. Mol. Sci. 17, 2143 (2016).
Shirabe, K., Bekki, Y., Gantumur, D., Araki, K., Ishii, N., Kuno, A. et al. Mac-2 binding protein glycan isomer (M2BPGi) is a new serum biomarker for assessing liver fibrosis: more than a biomarker of liver fibrosis. J. Gastroenterol. 53, 819–826 (2018).
Toyoda, H., Kumada, T., Tada, T., Kaneoka, Y., Maeda, A., Korenaga, M. et al. Serum WFA+ -M2BP levels as a prognostic factor in patients with early hepatocellular carcinoma undergoing curative resection. Liver Int. 36, 293–301 (2016).
Bekki, Y., Yoshizumi, T., Shimoda, S., Itoh, S., Harimoto, N., Ikegami, T. et al. Hepatic stellate cells secreting WFA(+) -M2BP: Its role in biological interactions with Kupffer cells. J. Gastroenterol. Hepatol. 32, 1387–1393 (2017).
Klover P. J., Thangapazham R. L., Kato J., Wang J. A., Anderson S. A., Hoffmann V. et al. Tsc2 disruption in mesenchymal progenitors results in tumors with vascular anomalies overexpressing Lgals3. Elife. 6, e23202 (2017).
Yoo, B. C., Hong, S. H., Ku, J. L., Kim, Y. H., Shin, Y. K., Jang, S. G. et al. Galectin-3 stabilizes heterogeneous nuclear ribonucleoprotein Q to maintain proliferation of human colon cancer cells. Cell Mol. Life Sci. 66, 350–364 (2009).
Jiang, S. S., Weng, D. S., Wang, Q. J., Pan, K., Zhang, Y. J., Li, Y. Q. et al. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J. Transl. Med. 12, 273 (2014).
K., H. D., DC, A., George, J. K. C., Jung‐Ta, C., Ri‐Yao, Y. & Fu‐Tong, L. Galectin‐3 expression is induced in cirrhotic liver and hepatocellular carcinoma. Int. J. Cancer 81, 519–526 (1999).
Matsuda, Y., Yamagiwa, Y., Fukushima, K., Ueno, Y. & Shimosegawa, T. Expression of galectin-3 involved in prognosis of patients with hepatocellular carcinoma. Hepatol. Res. 38, 1098–1111 (2008).
Tsukagoshi, M., Araki, K., Yokobori, T., Altan, B., Suzuki, H., Kubo, N. et al. Overexpression of karyopherin-α2 in cholangiocarcinoma correlates with poor prognosis and gemcitabine sensitivity via nuclear translocation of DNA repair proteins. Oncotarget 8, 42159–42172 (2017).
Luo, D., Yi, Y., Peng, K., Liu, T., Yang, J., Liu, S. et al. Oridonin derivatives as potential anticancer drug candidates triggering apoptosis through mitochondrial pathway in the liver cancer cells. Eur. J. Med Chem. 178, 365–379 (2019).
Guzelcan, E. A., Baxendale, I. R., Cetin-Atalay, R. & Baumann, M. Synthesis of new derivatives of boehmeriasin A and their biological evaluation in liver cancer. Eur. J. Med Chem. 166, 243–255 (2019).
Haberle, V., Forrest, A. R., Hayashizaki, Y., Carninci, P. & Lenhard, B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 43, e51 (2015).
Frith, M. C., Saunders, N. F., Kobe, B. & Bailey, T. L. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071 (2008).
Koths, K., Taylor, E., Halenbeck, R., Casipit, C. & Wang, A. Cloning and characterization of a human Mac-2-binding protein, a new member of the superfamily defined by the macrophage scavenger receptor cysteine-rich domain. J. Biol. Chem. 268, 14245–14249 (1993).
Narimatsu, H. Development of M2BPGi: a novel fibrosis serum glyco-biomarker for chronic hepatitis/cirrhosis diagnostics. Expert Rev. Proteomics 12, 683–693 (2015).
van Meer, S., van Erpecum, K. J., Sprengers, D., Coenraad, M. J., Klumpen, H. J., Jansen, P. L. et al. Hepatocellular carcinoma in cirrhotic versus noncirrhotic livers: results from a large cohort in the Netherlands. Eur. J. Gastroenterol. Hepatol. 28, 352–359 (2016).
Grassadonia, A., Tinari, N., Iurisci, I., Piccolo, E., Cumashi, A., Innominato, P. et al. 90K (Mac-2 BP) and galectins in tumor progression and metastasis. Glycoconj. J. 19, 551–556 (2002).
Inohara, H., Akahani, S., Koths, K. & Raz, A. Interactions between galectin-3 and Mac-2-binding protein mediate cell-cell adhesion. Cancer Res. 56, 4530–4534 (1996).
Wu, K. L., Huang, E. Y., Jhu, E. W., Huang, Y. H., Su, W. H., Chuang, P. C. et al. Overexpression of galectin-3 enhances migration of colon cancer cells related to activation of the K-Ras-Raf-Erk1/2 pathway. J. Gastroenterol. 48, 350–359 (2013).
Lin, L., Han, M. M., Wang, F., Xu, L. L., Yu, H. X. & Yang, P. Y. CXCR7 stimulates MAPK signaling to regulate hepatocellular carcinoma progression. Cell Death Dis. 5, e1488 (2014).
Bao, J., Walliander, M., Kovacs, F., Nagaraj, A. S., Hemmes, A., Sarhadi, V. K. et al. Spa-RQ: an image analysis tool to visualise and quantify spatial phenotypes applied to non-small cell lung cancer. Sci. Rep. 9, 17613 (2019).
Duvoux, C. & Toso, C. mTOR inhibitor therapy: Does it prevent HCC recurrence after liver transplantation? Transpl. Rev. (Orlando) 29, 168–174 (2015).
Koeberle, D., Dufour, J. F., Demeter, G., Li, Q., Ribi, K., Samaras, P. et al. Sorafenib with or without everolimus in patients with advanced hepatocellular carcinoma (HCC): a randomized multicenter, multinational phase II trial (SAKK 77/08 and SASL 29). Ann. Oncol. 27, 856–861 (2016).
Okuno T., Kakehashi A., Ishii N., Fujioka M., Gi M. & Wanibuchi H. mTOR Activation in liver tumors is associated with metabolic syndrome and non-alcoholic steatohepatitis in both mouse models and humans. Cancers (Basel). 10, 465 (2018).
Wang, C., Cigliano, A., Delogu, S., Armbruster, J., Dombrowski, F., Evert, M. et al. Functional crosstalk between AKT/mTOR and Ras/MAPK pathways in hepatocarcinogenesis: implications for the treatment of human liver cancer. Cell Cycle 12, 1999–2010 (2013).
Wang, S., Zhu, M., Wang, Q., Hou, Y., Li, L., Weng, H. et al. Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 9, 1027 (2018).
Zhu, A. X., Chen, D., He, W., Kanai, M., Voi, M., Chen, L. T. et al. Integrative biomarker analyses indicate etiological variations in hepatocellular carcinoma. J. Hepatol. 65, 296–304 (2016).
Acknowledgements
We would like to thank the Sysmex Corporation for providing M2BPGi. We are deeply grateful to the Otsuka Toshimi Scholarship Foundation for providing financial assistance. We warmly thank Dr Nakazawa Seshiru for his participation in this work. We would like to thank Enago (www.enago.jp) for the English language review.
Author information
Authors and Affiliations
Contributions
D.G. designed the study and wrote the initial draft of the paper. N.H., T.Yo., K.S., N.I., T.Ya., K.H., M.T., T.I., A.W., N.K. and K.A. contributed to analysis and data interpretation, and assisted in the preparation of the paper. T.H., T.O. and H.K. have contributed to data collection and interpretation, and have critically reviewed the paper. All authors have approved the final version of the paper and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The establishment of tissue sample from surgically resected HCC tissues was approved by the Institutional Review Board of Gunma University (Approval number: HS2018-280). Consent to participate was obtained from the patients. All mouse experiments were conducted in compliance with the guidelines of the Institute for Laboratory Animal Research at Gunma University, Maebashi, Japan. The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Data availability
All data generated or analysed during this study are included in this paper. CAGE data reported in this study have been deposited to the GEO (Gene Expression Omnibus) and are available through the accession number [GSE134471].
Competing interests
The authors declare no competing interests.
Funding information
This study was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS), grant numbers 17K19893, 18K07665 and 18H02877. The work was also supported in part by Research Grant of the Princess Takamatsu Cancer Research Fund, Suzuken Memorial Foundation and Pancreas Research Foundation of Japan.
Additional information
Note This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dolgormaa, G., Harimoto, N., Ishii, N. et al. Mac-2-binding protein glycan isomer enhances the aggressiveness of hepatocellular carcinoma by activating mTOR signaling. Br J Cancer 123, 1145–1153 (2020). https://doi.org/10.1038/s41416-020-0971-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-020-0971-y
This article is cited by
-
Cellular senescence-related genes: predicting prognosis in hepatocellular carcinoma
BMC Cancer (2023)
-
Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling
Cell Death & Disease (2022)
-
RAP2A promotes apoptosis resistance of hepatocellular carcinoma cells via the mTOR pathway
Clinical and Experimental Medicine (2021)