Abstract
Background
Metastatic colorectal cancer (mCRC) is a highly heterogeneous disease from a clinical, molecular, and immunological perspective. Current predictive models rely primarily in tissue based genetic analysis, which not always correlate with inflammatory response. Here we evaluated the role of a circulating inflammatory signature as a prognostic marker in mCRC.
Methods
Two hundred eleven newly diagnosed patients with mCRC were enrolled in the study. One hundred twenty-one patients had unresectable metastases, whereas ninety patients had potentially resectable liver metastases at presentation. Analysis of miR-21, IL-6, and IL-8 in the plasma of peripheral blood was performed at baseline. Patients with high circulating levels of ≥2 of the three inflammation markers (miR-21, IL-6, and IL-8) were considered to have the “Inflammation phenotype-positive CISIG”.
Results
Positive CISIG was found in 39/90 (43%) and 50/121 (45%) patients in the resectable and unresectable cohort, respectively. In the resectable population the median relapse-free survival was 18.4 compared to 31.4 months (p = 0.001 HR 2.09, 95% CI 1.2–3.67) for positive vs. negative CISIG. In contrast, the individual components were not significant. In the same population the median overall survival was 46.2 compared to 66.0 months (p = 0.0003, HR 2.57, 95% CI 1.26–5.27) for positive vs. negative CISIG, but not significant for the individual components. In the unresectable population, the median overall survival was 13.5 compared to 25.0 months (p = 0.0008, HR 2.49, 95% CI 1.46–4.22) for positive vs. negative CISIG. IL-6 was independently prognostic with overall survival of 16.2 compared to 27.0 months (p = 0.004, HR 1.96, 95% CI 1.24–3.11) for high vs. low IL-6, but not the other components. Using a Cox regression model, we demonstrated that CISIG is an independent predictive marker of survival in patients with unresectable disease (HR 1.8, 95% CI 1.2, 2.8, p < 0.01).
Conclusion
In two different cohorts, we demonstrated that CISIG is a strong prognostic factor of relapse-free and overall survival of patients with mCRC. Based on these data, analysis of circulating inflammatory signaling can be complimentary to traditional molecular testing.
Similar content being viewed by others
Background
Better understanding of colorectal cancer (CRC) biology led to the development of novel combination therapies that prolong patients’ survival.1 Nevertheless, approximately a quarter of patients with stage II and the majority of patients with stage III and IV disease require multiple lines of systemic therapy and eventually succumb from the disease.2 Identification of patients with increased risk of progression is critical to understand critical components of the disease biology, prioritize therapeutic interventions, and improve clinical outcomes.
The association of chronic inflammation and CRC tumorigenesis is well established and in the last decade has received growing supportive evidence from epidemiological, genetic, and pharmacological studies. The crosstalk between inflammatory, stromal, and cancer cells occurs in multiple levels allowing the tumor cells to undergo genetic, epigenetic, and biochemical changes to acquire an aggressive phenotype. MicroRNAs are a critical component of this cycle. miR-21 is specifically relevant to CRC tumorigenesis.3,4 Multiple studies have demonstrated that miR-21 is overexpressed in CRC human specimens, whereas the level of expression is higher in progressive stages of the disease.3,5,6,7,8 Animal studies demonstrated that miR-21 is associated with inflammation-induced CRC tumorigenesis.9,10 Using human samples, Schetter et al. demonstrated that the pro-inflammatory effect of miR-21 is mediated through cytokine upregulation including IL-6 and IL-8, two cytokines that have also been directly associated with CRC progression.11 Aberrant expression of miR-21 in tumor tissue is associated with reduced disease-free survival (DFS) and poor overall survival (OS) of patients with CRC, independently of clinical pathologic and major tumor genetic features.8,12,13 Further, Sarlinova et al. demonstrated that mirR-21 expression is significantly increased in the peripheral blood of patients with CRC, compared to matched healthy individuals.14 Based on this knowledge, three independent studies evaluated the role of serum miR-21 levels as a biomarker in CRC. In contrary to studies performed on tissue samples, serum levels of miR-21 alone have limited prognostic value.15
Tissue-based analyses have not always well reflected the current state of the tumor-host interactions. Indeed, cancer cachexia, systemic inflammatory response, and tumor-associated symptom burden are poorly evaluated by studies of the tumor tissue. Additionally, tumor-based assessments are limited to a single time-point in the disease course and do not fully reflect the current state of the disease. Circulating markers, in contrast, provide temporal relevance and integrate a systemic response to the tumor.
On the basis of this rationale, in this study, we examined the role of Circulating Inflammation SIGnature (CISIG) determined by high miR-21, IL-6, and IL-8 circulating levels as a potential marker in metastatic CRC. In two different cohorts, we demonstrated that CISIG is a strong prognostic factor of progression-free and overall survival of patients with metastatic CRC. Therefore, the inflammatory signature might be valuable tool in secondary prevention and patient selection for aggressive medical management.
Methods
Study cohorts
Two hundred eleven (211) newly diagnosed patients with metastatic colorectal cancer who presented to MD Anderson Cancer Center from April 2002 to December 2008 were enrolled in the study. The patients were divided into two cohorts based on clinical characteristics. The first cohort included one hundred twenty-one (121) patients with unresectable metastases at presentation, whereas the second validation cohort included ninety patients (90) with potentially resectable metastases at presentation. Patients from the second cohort subsequently underwent surgical intervention including partial hepatectomy, with curative intent within 2 years of the date of plasma draw. Patient characteristics are shown in Table 1. The study was carried out with the approval of MD Anderson Cancer Center Institutional Review Board, in accordance to the declaration of Helsinki.
Blood collection
Human plasma samples for CISIG analysis were obtained after informed consent had been obtained from the patient, per Institutional Review Board (IRB) protocol at the MD Anderson Cancer Center (MDACC). Samples were drawn at the time of presentation, and before initiation of any systematic therapy.
Patients follow-up
Progression-free survival (PFS) was defined as the interval between enrollment on the study and clinical, laboratory or radiologic progression, or death from any cause. Relapse-free survival (RFS) was defined as the interval between liver resection and recurrence in liver or elsewhere, or death from any cause. Overall survival (OS) was defined as the interval between enrollment on the study and death from any cause.
Sample collection and storage
Plasma samples were drawn and collected in ethylenediaminetetraacetic acid tubes and centrifuged at 2800 r.p.m. for 10 min at −22 °C. Sample aliquots were placed into 0.5-mL cryovials and stored at −70 °C to −80 °C until analysis. Plasma samples (in 500-μL aliquots) were thawed in parallel and each used for suspension bead multiplex assays.
MicroRNA isolation and quantitative RT-PCR
For microRNA-based RT-PCR assays, 2.5 µL of enriched small RNAs from plasma samples were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, San Diego, CA) according to manufacturer’s instructions in a total reaction volume of 7.5 µL. A 1∶20 dilution of RT products was used as template for the PCR stage. PCR reaction was performed in triplicate wells using TaqMan 2× Universal PCR Master Mix with conditions as described previously. No-template controls for both RT step and PCR step were included to ensure target specific amplification. The 7900 Sequence Detection System 2.3 (Applied Biosystems) software defaults were used to compute the relative change in RNA expression by the 2−ΔΔCt method with 95% confidence intervals. Circulating miR-21 levels were determined using levels of mir-16 as a control (normalizer). All hemolytic samples (evaluated by spectrophotometric method) were excluded from the study.
Cytokine assay
Levels of cytokines and biomarkers in serum were assessed using multiplex bead assay (Bio-Rad Laboratories, Hercules, CA, USA and EMD, Bioscience Research Reagents, Temecula, CA, USA). Briefly, samples were incubated for 1 h on the array plates that were pre-spotted with capture antibodies specific for each protein biomarker. Plates were decanted and washed four times before adding a cocktail of biotinylated detection antibodies to each well. After incubating with detection antibodies, plates were washed four times and incubated with streptavidin-horseradish peroxidase conjugate. All incubations were done for 30 min at room temperature with shaking at 200 rpm. Plates were again washed before adding a chemiluminescent substrate, followed by immediate imaging. Cytokine array data are available upon request to the corresponding author.
Statistical analysis
Statistical analysis was performed using STATA version 11.0. The cut-off of the highest tertile was used to define high level of circulating miR-21 and the bottom two tertiles were defined as low level, based on previous literature.3 For IL-6 and IL-8, the median was used as a cut-off point to dichotomize the levels of these markers to high vs. low. Patients with high circulating levels of >2 of the 3 inflammation markers (miR-21, IL-6, and IL-8) were considered to have the “Inflammation phenotype-positive CISIG”. Spearman correlation coefficients for correlation between circulating miR-21, IL-6, and IL-8 were computed. Median OS, and disease-free survival were estimated non-parametrically using the Kaplan–Meier method and compared by the log-rank test. Cox proportional hazards regression was used to adjust for potential confounders and significant differences were assessed using the log-rank test. Calculations were performed with SPSS-version 23.0 software (IBM Corp., Armonk, NY) and SAS 9.4 (SAS Institute). p values of less than 0.05 were considered statistically significant. The OS and corresponding censoring were computed in months from diagnosis to death for each patient. Additionally, progression-free survival and recurrent-free survival was computed for patients and was defined as the interval between blood draw and recurrent disease.
Results
Inflammation serum signature is prevalent in metastatic CRC
Circulating miR-21, IL-6, and IL-8 levels were measured at diagnosis of metastatic disease in both resectable and unresectable patient cohorts. Positive circulating inflammation signature (CISIG) was found in 39/90 (43%) and 50/121 (45%) patients in the resectable and unresectable cohort, respectively. The difference in CISIG prevalence between the two cohorts was not statistically significant (p > 0.05) (Table 1).
Correlations of plasma levels of inflammatory markers
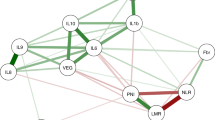
The correlation between inflammatory markers was performed using Spearman’s coefficient. In agreement with previous studies performed in CRC tissue samples, the serum levels of miR-21, IL-6, and IL-8 showed statistically significant correlation (p < 0.001). In the integrated study population (211 patients), the Spearman’s correlation coefficient between miR-21/ IL-6, miR-21/ IL-8, and IL-6/ IL-8 was 0.38, 0.38, and 0.76 respectively (data not shown).
Inflammatory signature predicts relapse-free survival and overall survival after hepatectomy
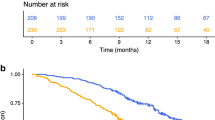
In the resectable population, we evaluated the correlation between inflammatory markers and inflammatory signature at the time of hepatectomy with relapse-free survival, as determined by the time between hepatectomy and relapse of the disease. The median relapse-free survival was 18.4 compared to 31.4 months (p = 0.001 HR 2.09, 95% CI 1.2–3.67) for positive vs. negative CISIG. In contrast, the individual components were not significant: 19.7 compared to 24.2 months (p = 0.24, HR 1.434, 95% CI 0.79–2.6) for high vs. low miR-21, 18.4 compared to 28.7 months (p = 0.26, HR 1.36, 95% CI 0.80–2.31) for high vs. low IL-6, and 21.7 compared to 24.2 months (p = 0.61, HR 1.23, 95% CI 0.72–2.80) for high vs. low IL-8, and (Fig. 1 and Supplementary Table 1). In this population, overall survival is further dictated by efficacy of treatment after relapse, and therefore overall survival is also relevant to incorporate risk of recurrence and treatment benefit. The median overall survival was 46.2 compared to 66.0 months (p = 0.0003, HR 2.57, 95% CI 1.26–5.27) for positive vs. negative CISIG, but not significant for the individual components: 46.2 compared to 66.0 months (p = 0.05, HR 2.1, 95% CI 1.01–4.46) for high vs. low miR-21, 46.2 compared to 66.4 months (p = 0.1, HR 1.7, 95% CI 0.90–3.30) for high vs. low IL-6, and 46.2 compared to 62.9 months (p = 0.24, HR 1.5, 95% CI 0.77–2.80) for high vs. low IL-8 (Fig. 2; Table 2).
Inflammatory signature predicts survival in of patients with unresectable disease
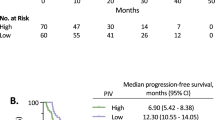
The role of individual inflammatory markers and the CISIG in predicting survival in patients with unresectable CRC was examined. The median overall survival in this population was 13.5 compared to 25.0 months (p = 0.0008, HR 2.49, 95% CI 1.46–4.22) for positive vs. negative CISIG. IL-6 was independently prognostic with overall survival of 16.2 compared to 27.0 months (p = 0.004, HR 1.96, 95% CI 1.24–3.11) for high vs. low IL-6, but not the other components: 19.7 compared to 24.2 months (p = 0.23, HR 1.43, 95% CI 0.79–2.61) for high vs. low miR-21, and 19.1 compared to 24.3 months (p = 0.11, HR 0.8, 95% CI 0.15–1.43) for high vs. low IL-8 (Fig. 3; Table 3).
Inflammatory signature is an independent predictive marker of survival in patients with unresectable disease
Previous studies have demonstrated that elevated CEA, low hemoglobin, elevated lymphocyte to neutrophil ratio (LNR), elevated WBC, elevated platelets, and previous chemotherapy are associated with poor prognosis in metastatic colorectal cancer.16,17 Using our unresected patient cohort, we evaluated the ability of the inflammatory signature to independently predict overall survival. We included variables significantly associated with the inflammatory signature, and known prognostic factors, such as patient sex, and site of the primary tumor. The final model included patient sex, primary tumor site (left vs. right), Hgb < 10 g/dL, CEA, NLR > 5, previous cytotoxic chemotherapy, albumin < 3.5 g/dL, platelets > 310 000 (cells/mcL), and WBC > 8000 (cells/mcL) as covariates. After adjusting for the above covariates, the inflammatory signature was still significantly predictive of overall survival (HR 1.8, 95% CI 1.2, 2.8, p < .01). The only other significant predictor in the model was left sided tumor (which was protective, HR: 0.6, 95% CI: 0.4, 0.9, p = 0.02) (Table 3). Finally, we examined the correlation of CISIG with other variables used in the model. Our analysis showed that in this patient cohort CISIG was associated with previous chemotherapy, albumin < 3.5, increased CEA, hemoglobin of <12.4, NLR > 5, platelet counts > 310 and WBC > 8. We run these seven factors through model selection for their ability to predict a positive CISIG and found that only albumin < 3.5, hemoglobin < 12.4, platelet counts > 310, and WBC > 8 remained significantly associated with positive CISIG in a multivariate model (Supplementary Table 2).
Discussion
Individual inflammatory markers and cytokines have previously shown prognostic value in solid tumors in small trials. In this study, we demonstrate that a signature of circulating inflammatory markers correlates with metastatic colorectal cancer aggressiveness. Of note, the prognostic value of the CISIG is evaluated in both patients with resectable and unresectable tumors with similar statistically significant results.
A number of studies demonstrated a correlation between genetic mutations and aggressive disease. The association of Ras mutations with poor disease outcomes after hepatectomy is well documented.18 In parallel, the prognostic impact of BRAFV600E mutations in metastatic CRC is also dramatic, since patients with BRAFV600E-mutant CRC survive approximately less than half as long as patients with BRAF wild-type (WTBRAF) metastatic disease.19 In contrary, BRAF mutations that occur outside the codon 600 define a clinically distinct phenotype of metastatic CRC with excellent prognosis.20 In a recent study, we demonstrated that the double mutation of APC and PIK3CA in tissue samples from liver metastases was associated with reduced RFS and OS rates, independently of Ras mutation status and surgical management.21 However, these features do not correlate well with the inflammatory response of the host to the tumor. As such, analysis of inflammatory signaling can be complimentary to traditional molecular testing.
The mechanism of miR-21, IL-6, and IL-8-induced CRC progression is not clearly understood. Studies performed in preclinical models showed that miR-21 downregulates tumor-suppressor genes including PTEN, and causes upregulation oncologic pathways such us RhoB.22,23,24,25,26,27 In parallel, activation of NF-kB/IL-6 paracrine axis promotes proliferation of CRC epithelial cells,28,29 whereas the IL-6-STAT3-mir-34a signaling pathway enhances the EMT phenotype.30 In agreement with the above findings, the Src/NF-kB/IL6 axis has been implicated in epithelial transformation in a positive feedback loop.31 Similarly, the IL-8-CXCR2 autocrine/paracrine axis has been implicated in proliferation of epithelial cells, EMT-phenotype, and VEGF-independent angiogenesis.32 Furthermore, it was proved, in a lung cancer model, that miR-21 can function as ligand to human Toll-like receptor (TLR) family, murine TLR8 in immune cells, triggering a TLR-mediated pro-metastatic inflammatory response including the secretion of cytokines such as TNF-α and IL-6, that ultimately may lead to tumor growth and metastasis.33
Poor outcomes and driving processes including EMT, stemness, and angiogenesis have been recapitulated in comprehensive gene expression studies such as the consensus molecular subtypes (CMS). The association of distinct CMSs with tumor inflammation and systemic inflammatory response is currently under evaluation. An initial report demonstrated that out of the four CMSs, two highly express immune-specific gene signatures. The good-prognosis microsatellite unstable (CMS1) is characterized by overexpression of genes specific to cytotoxic lymphocytes. In contrast, the poor-prognosis mesenchymal subgroup (CMS4) expresses markers of lymphocytes and of cells of monocytic origin.34 Based on these results, an inflammatory signature might be valuable in distinguishing CMSs and tailor therapy in patients with metastatic CRC.
MicroRNA may have a role in explaining groups defined by comprehensive gene expression in CRC. Network approaches combining transcription factor, methylation, and miRNA analysis demonstrate that up to 74.8% of variation seen in gene expression in mesenchymal vs. other subtypes is explained by miRNAs.35 Similarly, an alternate analysis comparing the most differentially expressed miRNAs to each CRC subtype demonstrated that these miRNAs also defined the mesenchymal phenotype.36 Both studies also found that downregulation of miR-200 (and family members) contributed significantly to regulating EMT, matrix remodeling, and TNF signaling via NF-kB. A master regulator function for miRNA is suggested by these findings and supports inclusion of miRNAs in future prognostic scores.
While this study demonstrates the potential applications of circulating inflammatory markers to determine aggressiveness of metastatic tumors and responses to treatment, it must be interpreted within the contexts of its limitations. Due to the methodology used, it lacks the external validity of cytokine assays owing to use of percentile cut-offs. Also, this study lacks the direct correlation of circulating markers with expression of the same markers in tissue samples from matched cases. Our assessment of inflammatory markers and clinical characteristics was blinded and samples were prospectively collected, both features that add to the validity of the experimental design but do not obviate the need for direct association of circulating markers with tissue marker expression.
In summary, we demonstrated that a three-marker circulating inflammation signature could be feasible and predictive of outcomes with standard systemic therapy and overall survival, and more importantly demonstrate the potential as a noninvasive assessment of the inflammatory state of the tumor.
References
Dienstmann, R. et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 17, 79–92 (2017).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. Cancer J. Clin. 66, 7–30 (2016).
Schetter, A. J. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299, 425–436 (2008).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA 103, 2257–2261 (2006).
Slaby, O. et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72, 397–402 (2007).
Nielsen, B. S. et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastas. 28, 27–38 (2011).
Yantiss, R. K. et al. Clinical, pathologic, and molecular features of early-onset colorectal carcinoma. Am. J. Surg. Pathol. 33, 572–582 (2009).
Mima, K. et al. MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival. Clin Cancer Res. 22, 3841–3848 (2016).
Shi, C. et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 65, 1470–1481 (2016).
Iliopoulos, D., Jaeger, S. A., Hirsch, H. A., Bulyk, M. L. & Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 39, 493–506 (2010).
Schetter, A. J. et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin. Cancer Res. 15, 5878–5887 (2009).
Kjaer-Frifeldt, S. et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br. J. Cancer 107, 1169–1174 (2012).
Oue, N. et al. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int. J. Cancer 134, 1926–1934 (2014).
Sarlinova, M. et al. miR-21, miR-221 and miR-150 are deregulated in peripheral blood of patients with colorectal cancer. Anticancer Res. 36, 5449–5454 (2016).
Conev, N. V. et al. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci. Trends 9, 393–401 (2015).
Sjoquist, K. M. et al. Personalizing survival predictions in advanced colorectal cancer: The ARCAD Nomogram Project. J. Natl Cancer Inst. 110, 638–648 (2018).
Dolan, R. D., McSorley, S. T., Horgan, P. G., Laird, B. & McMillan, D. C. The role of the systemic inflammatory response in predicting outcomes in patients with advanced inoperable cancer: systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 116, 134–146 (2017).
Vauthey, J. N. et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann. Surg. 258, 619–626 (2013).
Korphaisarn, K. & Kopetz, S. BRAF-directed therapy in metastatic colorectal cancer. Cancer J. 22, 175–178 (2016).
Jones, J. C. et al. Non-V600 BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J. Clin. Oncol. 35, 2624–2630 (2017).
Chun Y. S. et al. Deleterious effect of RAS and evolutionary high-risk TP53 double mutation in colorectal liver metastases. Ann. Surg. 2017.
Selaru, F. M. et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology 49, 1595–1601 (2009).
Wang, Z. X., Lu, B. B., Wang, H., Cheng, Z. X. & Yin, Y. M. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch. Med. Res. 42, 281–290 (2011).
Zhu, S., Si, M. L., Wu, H. & Mo, Y. Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336 (2007).
Zhu, S. et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359 (2008).
Ziyan, W., Shuhua, Y., Xiufang, W. & Xiaoyun, L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med. Oncol. 28, 1469–1474 (2011).
Liu, M. et al. miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett. 585, 2998–3005 (2011).
Grivennikov, S. et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113 (2009).
Bollrath, J. et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91–102 (2009).
Rokavec, M. et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 124, 1853–1867 (2014).
Iliopoulos, D., Hirsch, H. A. & Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009).
Fernando, R. I., Castillo, M. D., Litzinger, M., Hamilton, D. H. & Palena, C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 71, 5296–5306 (2011).
Fabbri, M. et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. USA 109, E2110–E2116 (2012).
Becht, E. et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin. Cancer Res. 22, 4057–4066 (2016).
Fessler, E. et al. A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene 35, 6026–6037 (2016).
Cantini, L. et al. MicroRNA-mRNA interactions underlying colorectal cancer molecular subtypes. Nat. Commun. 6, 8878 (2015).
Acknowledgements
Work in Dr. Calin’s laboratory is supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC), the NIH/NCI grant 1 R01 CA182905-01, a U54 grant—UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project, a Team DOD (CA160445P1) grant, a Ladies Leukemia League grant, a CLL Moonshot Flagship project, a SINF 2017 grant, and the Estate of C. G. Johnson, Jr. Work in Dr. Kopetz’s laboratory is supported by NIH grants R01 CA184843, R01 CA172670, R01 CA187238, and a CRC Moonshot project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Note:
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution 4.0 International (CC BY 4.0).
Supplementary information
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varkaris, A., Katsiampoura, A., Davis, J.S. et al. Circulating inflammation signature predicts overall survival and relapse-free survival in metastatic colorectal cancer. Br J Cancer 120, 340–345 (2019). https://doi.org/10.1038/s41416-018-0360-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-018-0360-y
This article is cited by
-
Which patients are prone to suffer liver metastasis? A review of risk factors of metachronous liver metastasis of colorectal cancer
European Journal of Medical Research (2022)
-
A pseudo-values regression model for non-fatal event free survival in the presence of semi-competing risks
Statistical Methods & Applications (2022)
-
Modeling of the immune response in the pathogenesis of solid tumors and its prognostic significance
Cellular Oncology (2020)