Abstract
We sought to define the genomic landscape of diffuse large B-cell lymphoma (DLBCL) by using formalin-fixed paraffin-embedded (FFPE) biopsy specimens. We used targeted sequencing of genes altered in hematologic malignancies, including DNA coding sequence for 405 genes, noncoding sequence for 31 genes, and RNA coding sequence for 265 genes (FoundationOne-Heme). Short variants, rearrangements, and copy number alterations were determined. We studied 198 samples (114 de novo, 58 previously treated, and 26 large-cell transformation from follicular lymphoma). Median number of GAs per case was 6, with 97% of patients harboring at least one alteration. Recurrent GAs were detected in genes with established roles in DLBCL pathogenesis (e.g. MYD88, CREBBP, CD79B, EZH2), as well as notable differences compared to prior studies such as inactivating mutations in TET2 (5%). Less common GAs identified potential targets for approved or investigational therapies, including BRAF, CD274 (PD-L1), IDH2, and JAK1/2. TP53 mutations were more frequently observed in relapsed/refractory DLBCL, and predicted for lack of response to first-line chemotherapy, identifying a subset of patients that could be prioritized for novel therapies. Overall, 90% (n = 169) of the patients harbored a GA which could be explored for therapeutic intervention, with 54% (n = 107) harboring more than one putative target.
Key points
-
This study demonstrates the applicability of CLIA-compliant targeted FFPE sequencing for large-scale clinical trials.
-
TP53mut is the main predictor of refractoriness or early relapse.
Similar content being viewed by others
Introduction
The ability to identify and therapeutically target patient-specific genomic alterations has made precision oncology a reality for several types of cancer1. Unfortunately, in aggressive lymphomas, no approved genomic biomarker-driven therapies are standard of care. The problem is exemplified by diffuse large B-cell lymphoma (DLBCL) where, despite a relapse rate of over 30%, RCHOP is being administered as an almost uniform first-line of care, over two decades since it was introduced2. Thus, there is an unmet need to develop genomic biomarker-driven therapeutics, to improve outcomes for patients with DLBCL.
Next-generation sequencing studies have produced a vast array of data regarding the underlying genomic alterations (GAs) that characterize DLBCL. These demonstrate a striking genetic heterogeneity that likely accounts for the observed variability in clinical phenotype3,4,5,6,7. Recurrent alterations have been identified in over 300 genes, none of which pathognomonic, as all occur at frequencies <30%, usually <10%7. Identifying the clinical implications of these alterations requires large cohorts, and the integration of several testing modalities (e.g. DNA sequencing to identify short nucleotide variations (SNVs) and copy number alterations (CNAs), and RNA sequencing for gene rearrangements)8. In this regard, most genomic studies in DLBCL have been carried out in the research setting, often implementing assays such as whole genome or whole exome sequencing using fresh frozen biopsy specimens7. These may be prohibitively resource intensive for adaptation in large-scale clinical trials or in everyday practice. In these settings, integrated hybridization capture of both DNA and RNA using formalin-fixed paraffin-embedded (FFPE) specimens may be most appropriate7,9. Potential advantages of this approach include: (1) simultaneous detection of all classes of GAs including short variants (base substitutions and small indels), CNAs, and gene rearrangements/fusions; (2) sensitive detection of fusion transcripts involving the genes of interest due to inclusion of the RNA sequencing component; (3) flexibility in sample acquisition, storage and transportation when using FFPE; (4) high depth of sequencing coverage to enhance detection of rare variants even in samples with extensive non-malignant stromal and immune cell contamination; (5) streamlined bioinformatics; and (6) compliance with The Clinical Laboratory Improvement Amendments (CLIA) standard. However, it remains to be established what spectrum of GAs will be observed in clinical FFPE DLBCL specimens, how specific GAs will correlate with clinical and pathologic phenotype and with patient outcomes, and how this information can be incorporated in clinical care.
Herein, we describe the application of commercially available, CLIA-compliant, integrated DNA and RNA targeted sequencing panel (FoundationOne-Heme) to a retrospective cohort of 198 FFPE DLBCL specimens for the identification of GAs with potential clinical significance.
Materials and methods
Study population
Archived FFPE biopsy specimens from 198 patients with DLBCL were obtained with approval from the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board. Biopsies were collected between 1989 and 2012. Inclusion criteria were histologically confirmed DLBCL with appropriate patient consent to perform genomic sequencing. Germinal center B-cell-like (GCB) or non-GCB cell-of-origin (COO) was assessed by immunohistochemistry (IHC) according to the Hans algorithm8. Baseline demographics and survival data were extracted from the clinical record.
Sample preparation and sequencing
Samples were sequenced using the FoundationOne-Heme platform that uses DNA sequencing to interrogate the entire coding sequence of 406 genes, selected introns of 31 genes involved in rearrangements, and utilizes RNA sequencing to interrogate 265 genes known to be somatically altered in human hematologic malignancies10. Detailed protocols for DNA and RNA extraction, cDNA synthesis, library construction, and hybrid selection as well as a survey of methodological validation tests, have been recently published, and are detailed in the supplementary methods and supplementary table 1 10,11. In brief, specimens were reviewed by a pathologist to confirm ≥20% tumor nuclei and a tissue volume of ≥2 mm3. We used 20% tumor content as the minimal requirement, having demonstrated in a previous validation study that the pipeline approaches a sensitivity of 100% for SNVs and CNAs above this cutoff10. Genomic DNA and RNA were extracted and fragmented to ~200 bp fragment size. Samples were tested to ensure sufficient DNA yield (50–200 ng) and RNA yield (≥3.5 ng/µL). Sequencing was performed with the Illumina HiSeq2500 system using 49 × 49 paired-end reads. Resultant sequences were analyzed for single nucleotide variants (SNVs—base substitutions and small indels), CNAs and rearrangements. Samples with median coverage <150× were considered failed and excluded from analysis. Known germline variants (per 1000 Genomes Project) were removed10,11. Significant non-synonymous variants were defined as any somatic alteration annotated in the COSMIC database (v62), as well as clear inactivating mutations (i.e. truncations or deletions) in established tumor suppressor genes10,11. The mutant allele frequency cutoff used for known somatic variants was 1%; 5% for potential driver somatic variants; 3% for previously described indels; and 10% for potential driver indels. Gene amplifications/gains were defined at a copy number ≥6, and gene losses as copy number of 0 10,11. For rearrangement identification we required a minimum of ten chimera reads for known fusions and 50 for potential driver rearrangements. Any aberration not meeting the aforementioned criteria was defined as Unknown Significance (UKS). Sequencing data are publicly available in an interactive format through the cBioPortal (www.cbioportal.org).
Individual genes were grouped together by the biologic pathways in which they operate (supplementary table 2)12,13. Actionable variants and pathways were defined by the presence of GAs predictive of response to an FDA-approved drug or an experimental agent in clinical trial14. These data were derived by a review of literature and publicly accessible databases including OncoKB, the FDA Pharmacogenomic Biomarkers in Drug Labeling, GeneCards, and clinicaltrials.gov12,13,14,15,16,17. Variant actionability was graded based on the OncoKB criteria. Level 1 was defined as alterations recognized by the FDA as predictive of response to an approved drug in DLBCL. Level 2 included non-FDA predictive biomarkers for response in DLBCL (2A) or FDA-approved biomarkers for response in a different malignancy (2B). Level 3 includes alterations supported by compelling data from clinical trials in DLBCL (3A) or another malignancy (3B). Level 4 are candidate biomarkers for response based on early clinical or preclinical studies (supplementary excel file 2)14.
Statistical analysis
Descriptive statistics are provided for all genes dichotomously (i.e. presence/absence of any alteration). Differences in alteration frequency between groups were determined using Fischer’s exact test, with differences in the total number of alterations across groups assessed using a Wilcoxon rank-sum test. Clustering analysis was done based on the Jaccard distance using the Ward D method. Analyses for response to treatment and survival were performed in the subset of patients with de novo disease treated with RCHOP (rituximab, cyclophosphamide, adriamycin, vincristine, steroids) or RCHOP-like chemotherapy. For the purpose of these analyses, tFL not previously treated was included with the de novo group, reasoning that in clinical practice the distinction between tFL at first diagnosis and DLBCL is not made easily, such that both conditions are treated similarly as de novo DLBCL and have comparable outcomes18. Median follow-up was estimated using the reverse Kaplan−Meier method. Overall and progression-free survival (OS/PFS) were defined as the time from initiation of frontline treatment until death of any cause or disease progression or death (for PFS), censoring at the end of follow-up. Differences in OS and PFS between groups were assessed using the Kaplan−Meier method as well as univariate Cox proportional hazards regression models. Where applicable we adjusted for false discovery (FDR) using the Benjamini−Hochberg approach. Analyses were done in R 3.4.0 (R foundation, Austria).
Results
Of 219 FFPE DLBCL samples attempted, 214 were successfully sequenced, indicating a success rate of 98%. Sixteen cases were excluded from the analysis (for inadequate clinical data, primary central nervous system lymphoma or large-cell transformation from indolent lymphomas other than FL), leaving 198 cases for this analysis: 114 cases were from newly diagnosed untreated patients (de novo), 58 from previously treated patients, and 26 from tFL cases. Cell of origin was determined in 177 cases, with 48% (n = 95) classified as GCB and 41% (n = 82) as non-GCB. Of the 114 patients sequenced at diagnosis, 30% (n = 35) were refractory to first-line treatment or subsequently relapsed during follow-up. The median unique sequencing coverage was 555 × [476–656] for DNA and median total pairs for RNA were >20×106.
Genomic alterations and pathways landscape
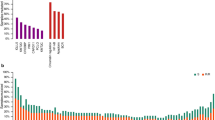
The median number of GAs per case was 6, with 97% of patients harboring at least one alteration (Table 1, supplementary figure 1 and supplementary excel file 1). The most commonly identified SNVs were in KMT2D (MLL2; 31%, n = 62), TP53 (24%, n = 48), MYD88 (18%, n = 36), CREBBP (18%, n = 35), and B2M (Beta-2-microglobulin; 17%; n = 33) (Fig. 1, Table 1). CNAs were identified in 42% (n = 84) of cases, involving 37 different genes. The most frequently identified losses were CDKN2A and/or CDKN2B (20% combined, n = 40), while the most frequent gene amplifications were observed in REL (8%, n = 16), CD274 (3%; n = 6), and MCL1 (3%, n = 6). Rearrangements (trans) were detected in 57% (n = 112) of cases involving 61 different genes. As expected, most involved the translocation of BCL2, BCL6, or MYC to the immunoglobulin heavy chain (IGH) enhancer (supplementary table 3). Deletion of CDKN2B was always accompanied by deletion of CDKN2A (though not vice versa) and associated with CD79Bmut, MYD88mut, PIM1mut, and PRDM1mut (Fig. 2, supplementary figure 2). CDKN2Bdel and CDKN2Adel were mutually exclusive with TP53mut (p < 0.001) as were BCL10mut (p = 0.04) and CD58mut (p = 0.04). A cluster of BCL2trans and KMT2Dmut corresponded with a GCB subtype and with high rates of TP53mut, EZH2mut, and TNFRSF14mut (p = 0.002; Fig. 2). Of note, the largest cluster of 80 patients (40%) did not have a distinct genomic signature.
Cluster analysis based on co-occurrence/anti-co-occurrence distance (“Jaccard”) with annotated pathways, R/R status (irrespective of time of sequencing) and cell of origin (by IHC). Presented are only genomic abnormalities present in ≥5% of the cohort. Dark-green/light-green (main plot)—presence/absence of a genomic abnormality respectively
Patients with R/R disease and tFL had higher rates of TP53mut compared to de novo patients (16% n = 18 de novo, 33% n = 19 R/R; and 42% n = 11 tFL; p = 0.02 R/R vs. de novo), and of KMT2Dmut (19% n = 22 de novo, 38% n = 22 R/R; and 69% n = 18 tFL; p = 0.01 R/R vs. de novo) (Table 1). Further, R/R and tFL cases were enriched for translocations (45% n = 51 de novo, 71% n = 41 R/R; and 77% n = 20 tFL; p = 0.002 for R/R vs. de novo). These were mainly IGH:BCL2 rearrangements for tFL (75% of cases), while among R/R patients 37% (n = 15) had a BCL2trans, 27% (n = 11) had a BCL6trans, and 12% (n = 5) had a MYCtrans.
These observations corresponded to a trend towards higher overall rates of abnormalities in tumor suppressors pathways that includes TP53mut and CDKN2Bdel (54% n = 61 de novo, 76% n = 44 R/R, and 69% n = 18 tFL, p = 0.007) and in the epigenetic histone modification pathway which includes KMT2Dmut (47% n = 41 non-relapsing, 64% n = 37 R/R, and 89% n = 23 tFL, p = 0.008) (Table 2). Of note, after correction for false discovery, none of these differences remained statistically significant. As expected, BCL2trans were more common in GCB compared to non-GCB (40% n = 38 vs. 5% n = 4, p < 0.001) as were CREBBPmut (27% n = 26 vs. 6% n = 5, p < 0.001), KMT2Dmut (43% n = 41 vs. 21% n = 17, p = 0.003), and TNFRSF14 (17% n = 16 vs. 4% n = 3, p = 0.01). CD79Bmut were observed solely in non-GCB (0 vs. 16% n = 13, p < 0.001), as was an enrichment for BCL6trans (11% n = 10 vs. 27% n = 22, p = 0.01) (supplementary table 4 and Fig. 3). We further observed an enrichment in MYD88mut, ETV6mut, and PRDM1mut among non-GCB and EZH2mut among GCB tumors; however, these did not remain significant after correction for FDR (supplementary table 5).
The median number of involved pathways was 4 with 90% of patients having at least two affected pathways (Table 2, Fig. 2). Overall, 90% (n = 179) of the patients harbored a GA which could be explored for therapeutic intervention, with 54% (n = 107) harboring more than one potential target. GAs involving a gene that is targeted by a drug approved by the FDA for another indication (level 2B) were identified in 56% (n = 110). These were comprised mainly of BCL2 inhibitors for BCL2-associated GAs, BTK inhibitors for MYD88mut, BRAF inhibitors for BRAFmut and immune check-point inhibitors in CD274 and PDCD1LG2 GAs (supplementary excel file 2). In 41% (n = 81) there was a GA targeted by a non-FDA-approved drug with compelling clinical evidence either in DLBCL (level 3A; 33%, n = 66; mostly histone deacetylase and EZH2 inhibitors in CREBBPmut, EP300mut, and EZH2mut) or in another indication (level 3B; 8%, n = 15). Finally, in 85% there was at least one target for a drug in preclinical or early clinical development (level 4).
Association of GAs with clinical presentations and pathologic subtypes
There were 106 de novo DLBCL patients treated with an RCHOP-based chemotherapy (58% RCOHP, 36% RCHOP-ICE, 7% DA-EPOCH-R). Median age was 58 (range 21–84) with a 61% male predominance. The majority (75%) had a stage III−IV disease, and 35% had an intermediate-high to high IPI. Complete response was observed in 88% (PR in 3% and SD/PD in 9%) of the patients and was not associated with stage, IPI, or other demographics (Table 3). Median follow-up was 66 months (95% CI 57–73) with 18 deaths and 26 disease progressions documented during that time, representing a 5y OS of 84% (95%CI 77–92%).
We investigated whether specific GAs were associated with patient outcomes, including response to chemotherapy or OS. We found that TP53 alterations predicted for lack of response to chemotherapy. Of the 12 patients with primary refractory disease, 8 (67%) were TP53mut. Of the 19 patients harboring a TP53mut, 10 (53%) did not achieve a complete response (CR) or relapsed within the first year, while one patient relapsed after 3 years. Six were without evidence of disease during a 2–5-year-follow-up, and two were still in remission at 12 and 22 months. TP53mut was also associated with shorter OS (5yOS 61% vs. 89% HR 5.8 95%CI 2.1–16, p = 0.001). Finally, TP53mut were detected in 19 R/R patients, of whom 13 (66%) were either refractory to their previous frontline therapy or had relapsed within less than a year.
Two other GAs were marginally associated with survival. B2Mmut (19% of patients; OS HR 2.9, 95%CI 1.1–7.6, p = 0.03), and CDKN2Adel when accompanied by CDKN2Bdel but not alone (14% of patients, OS HR 2.5 95%CI 0.9–7.1, p = 0.08). (Table 4, Fig. 3). In addition, abnormalities grouped under the protein translation machinery pathway (e.g. MYC, EIF4A2), present in 9% of patients, were associated with a shorter OS (HR 4.9 95%CI 1.7–13.8, p = 0.003), as were abnormalities in tumor suppressor pathways (which includes TP53mut). Lastly, patients with no alteration in TP53, CDKN2B or B2M, had a remarkably long OS (5yOS 96% vs. 74% for B2Mmut/CDKN2Bdel p = 0.03, and 60% for TP53mut p < 0.0001) (Fig. 3).
Discussion
Clinical trials that select patients for novel targeted therapies based on GAs require large-scale standardized sequencing endeavors, which can be facilitated by targeted DNA and RNA sequencing of FFPE specimens. This work describes the application of a CLIA-compliant integrated DNA and RNA targeted sequencing panel to a retrospective cohort of 198 FFPE DLBCL specimens for the identification of GAs with potential clinical significance.
Prior studies defining the genomic landscape of DLBCL have produced highly variable results2. For example, reported frequencies of CREBBP/EP300 mutations in DLBCL have ranged widely from 5 to 44%3,4,5,6,7. We detected alterations in the histone acetyltransferases CREBBP/EP300 in 21% of clinical FFPE DLBCL specimens. This frequency corresponds to the approximate 20% response rates to HDAC inhibitors observed in unselected patients with DLBCL, providing a biologic rationale for a clinical trial using CREBBP/EP300 mutation as a genomic biomarker to select DLBCL patients for treatment with the HDAC inhibitors (NCT02282358)19. Likewise, prior studies reported EZH2 mutations frequencies as high as 24%20, whereas we found EZH2mut in 11% of our cohort, a difference that would have major implications for designing a trial with sequencing-based selection of patients for treatment with EZH2 inhibitors.
Comprehensive genomic profiling also detected uncommon GAs (occurring at <3% frequency) with potential therapeutic relevance, which could help identify patients for genetically defined basket trials or select molecularly targeted therapies (supplementary excel file 2). For example, BRAF V600E and L597R/V mutations which may predict for response to BRAF or MEK inhibitors21. Likewise, we identified mutations in ID3 (L70P, P98R, Q100P) and TCF3 (N551K), which had been previously described, predominantly in Burkitt lymphoma, to confer dependence on PI3K pathway signaling, and may indicate a potential therapeutic vulnerability22.
In our study, 90% of the patients had at least one potentially targetable GA which could guide selection for clinical trials. Nearly two-thirds of these patients harbored two or more such targets. When using genomic profiling to define genomic biomarkers, it will be important to determine how variant allele frequencies (VAFs) of actionable mutations impact responses to molecularly targeted therapies. For example, we found that gain-of-function mutations in MYD88 L273P (a.k.a. L265P) exhibited VAFs that ranged from 9 to 74% (supplementary table 3). Thus, if the MYD88 mutations were targeted with a downstream IRAK1/4 inhibitor, the question arises as to whether tumors showing subclonal GAs with low VAFs will respond similarly to those with a high VAF dominant clone23. Although the impact of clonal and subclonal GAs on response to targeted therapies will become clearer as empiric observations are gathered, consensus regarding this question will need to be established soon, in order to design clinical trials that prospectively select patients based on genomic biomarkers16.
Consistent with previous reports, using non-clinical genome sequencing assays in the research setting, we confirmed that KMT2Dmut to be the most common SNV, present in 26% of de novo cases3,4,5,7,24,25. Similarly, TP53mut was noted in 16% of de novo disease and was enriched among R/R patients (33%)3,4,5,24,25. Further, in agreement with previous literature, certain GAs were highly associated with cell of origin designation, highlighting the need to account for molecular case-mix when comparing results from different genomic profiling studies. Compared to non-GCB, GCB samples were enriched for BCL2trans, CREBBPmut, KMT2Dmut, TNFRSF14mut, and EZH2mut, in keeping with rates described previously in the literature6,7,20,24. Likewise, non-GCB samples were enriched for CD79Bmut, BCL6trans, MYD88mut, ETV6mut, and PRDM1mut also at similar rates as previously described6,7,20,24.
Despite an accumulating body of research into the genomic landscape of DLBCL, very few GAs have been found to be associated with treatment refractoriness or disease relapse. Our study confirms prior associations between TP53mut and survival26,27. Though marginally significant, CDKN2A/Bdel and B2Mmut were also found to be associated with shorter OS28,29,30.
As larger sequencing cohorts are assembled, future studies will continue to refine the association between GAs and treatment outcomes. While many large centers use their own home-grown sequencing assays to select patients for targeted therapy, the use of a commercially available clinical assay can facilitate the application of precision medicine at local hospitals and doctor offices, in addition to facilitating the conduct of clinical trials across several institutions. Furthermore, the use of a standardized commercial assay may allow comparing the results of different clinical trials that select patients based on specific genetic alterations. Typically, the cost of a commercial assay is higher than institutional platforms. However, only large institutions are able to develop their own sequencing assays, leaving smaller centers to depend on third-party commercial vendors.
References
Garraway, L. A. Genomics-driven oncology: framework for an emerging paradigm. J. Clin. Oncol. 31, 1806–1814 (2013).
Roschewski, M., Staudt, L. M. & Wilson, W. H. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat. Rev. Clin. Oncol. 11, 12–23 (2014).
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 109, 3879–3884 (2012).
Morin, R. D. et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Zhang, J. et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 110, 1398–1403 (2013).
Reddy, A. et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 171, 481–494.e15 (2017).
Dubois, S.., & Jardin, F.. The role of next-generation sequencing in understanding the genomic basis of diffuse large B cell lymphoma and advancing targeted therapies. Expert Rev. Hematol. 4086, 1–15 (2016).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127 (24), 3004-14 (2016).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Younes, A. et al. The landscape of new drugs in lymphoma. Nat. Rev. Clin. Oncol. 14, 335–346 (2017).
Intlekofer, A. M. & Younes, A. Precision therapy for lymphoma? Current state and future directions. Nat. Rev. Clin. Oncol. 11, 585–596 (2014).
Chakravarty D., Gao J., Phillips S. & Kundra R. OncoKB: a precision oncology knowledge base. Precis Oncol. 2017;[epub].
Genomics—Table of Pharmacogenomic Biomarkers in Drug Labeling. https://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm. Accessed September 1, 2017.
Joffe, E., Iasonos, A. & Younes, A. Clinical trials in the genomic era. J. Clin. Oncol. 35, 1011–1017 (2017).
GeneCards—Human Genes Database. http://www.genecards.org/. Accessed September 23, 2017.
Guirguis, H. R. et al. Survival of patients with transformed lymphoma in the rituximab era. Ann. Hematol. 93, 1007–1014 (2014).
Lane, A. A. & Chabner, B. A. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 27, 5459–5468 (2009).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Holderfield, M., Deuker, M. M., McCormick, F. & McMahon, M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14, 455–467 (2014).
Schmitz, R. et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012).
Ngo, V. N. et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 (2011).
Dubois, S. et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin. Cancer Res. 22, 2919–2928 (2016).
Melchardt, T. et al. Clonal evolution in relapsed and refractory diffuse large B-cell lymphoma is characterized by high dynamics of subclones. Oncotarget 5, 51494–51502 (2016).
Xu-Monette, Z. Y. et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with rituximab-CHOP: a report from an International DLBCL Rituximab-CHOP Consortium Program study. Blood 120, 3986–3997 (2012).
Young, K.H., Leroy, K., & Moller, M.B. et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 112, 3088–3098 (2008).
Jardin, F. et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood 116, 1092–1104 (2010).
Challa-Malladi, M. et al. Combined genetic inactivation of β2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 20, 728–740 (2011).
Barrans, S. et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J. Clin. Oncol. 28, 3360–3365 (2010).
Acknowledgements
We thank the patients at Memorial Sloan Kettering Cancer Center who generously donated their biopsy specimens for research. We thank members of the Lymphoma Service at Memorial Sloan Kettering Cancer Center for helpful discussions and feedback. A.M.I. is supported by the NIH/NCI (K08 CA201483), Leukemia & Lymphoma Society (Special Fellow Award 3356-16), Burroughs Welcome Fund (Career Award for Medical Scientists 1015584), Susan and Peter Solomon Divisional Genomics Program, and Steven A. Greenberg Fund. The work was supported in part by the John and Barbara Vogelstein Foundation, the Memorial Sloan Kettering Cancer Center Support Grant NIH P30 CA008748, and the MSK SPORE in lymphoma (P50 CA192937). We thank Foundation Medicine, Inc., for performing the targeted sequencing assays as a collaborative research effort.
Author information
Authors and Affiliations
Contributions
A.M.I., E.J., and A.Y. conceived the project, analyzed the data, and wrote the manuscript. A.M.I., R.L.L., A.D.Z., A.D., M.L.P., M.R.v.d.B., and A.Y. helped design the content for the sequencing assay. J.H., M.K.N., S.E.T., T.M., and V.A.M. constructed and validated the sequencing assay, processed the samples, and performed the sequencing. P.H., E.J., and V.E.S. performed the statistical analysis. E.P. and M.M. assisted with bioinformatics analyses. A.R.C., A.D., C.L.B., C.H.M., C.P., D.J.S., M.M., A.N., S.H., J.F.G., A.M., P.H., A.K., K.M.K., and J.D.P. assisted with logistics of sample identification/collection, assessment of clinical data, and pathologic review.
Corresponding author
Ethics declarations
Conflict of interest
A.M.I., R.L.L., A.D.Z., A.D., M.L.P., M.R.v.d.B., and A.Y. have served as consultants for Foundation Medicine, Inc. J.H., M.K.N., S.E.T., and V.A.M. are employees of Foundation Medicine, Inc. V.A.M. has ownership interest (including patents) in Foundation Medicine, Inc. The remaining authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Intlekofer, A.M., Joffe, E., Batlevi, C.L. et al. Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay. Blood Cancer Journal 8, 60 (2018). https://doi.org/10.1038/s41408-018-0089-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41408-018-0089-0