Abstract
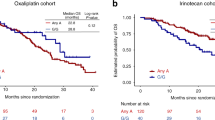
PIN1-mediated substrate isomerization plays a role in the repair of DNA double-strand breaks. We hypothesized that genetic polymorphisms in PIN1-related pathways may affect tumor sensitivity to oxaliplatin or irinotecan in metastatic colorectal cancer (mCRC) patients. We analyzed genomic DNA from five cohorts of mCRC patients (total 950) treated with different first-line treatments: oxaliplatin cohorts 1 (n = 146) and 2 (n = 70); irinotecan cohorts 1 (n = 228), and 2 (n = 276); and combination cohort (n = 230). Single nucleotide polymorphisms of candidate genes were analyzed by PCR-based direct sequencing. In the oxaliplatin cohort 1, patients carrying any PIN1 rs2233678 C allele had shorter progression-free survival (PFS) and overall survival (OS) than the G/G variant (PFS, 7.4 vs. 15.0 months, hazard ratio [HR] 3.24, P < 0.001; OS, 16.9 vs. 31.5 months, HR: 2.38, P = 0.003). In contrast, patients with C allele had longer median PFS than patients with G/G (11.9 vs. 9.4 months, HR: 0.64, 95%CI: 0.45–0.91, P = 0.009) in the irinotecan cohort 1. No significant differences were observed in the combination cohort. In comparison between the irinotecan cohort 1 and combination cohort, the patients carrying the G/G variant benefit greatly from the combination compared with irinotecan-based regimen (PFS, 11.6 vs. 9.4 months, HR 0.61, 95%CI: 0.47–0.78, P < 0.001; OS, 30.6 vs. 24.0 months, HR 0.79, 95%CI: 0.62–1.02, P = 0.060), while no significant difference was shown in any C allele. Germline PIN1 polymorphisms may predict clinical outcomes in mCRC patients receiving oxaliplatin-based or irinotecan-based therapy, and identify specific populations favorable to oxaliplatin plus irinotecan combination therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Govindaraghavan M, Lad AA, Osmani SA. The NIMA kinase is required to execute stage-specific mitotic functions after initiation of mitosis. Eukaryot Cell. 2014;13:99–109.
O’regan L, Blot J, Fry AM. Mitotic regulation by NIMA-related kinases. Cell Div. 2007;2:25.
Lu KP, Hunter T. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell. 1995;81:413–24.
Osmani AH, O’Donnell K, Pu RT, Osmani SA. Activation of the nimA protein kinase plays a unique role during mitosis that cannot be bypassed by absence of the bimE checkpoint. EMBO J. 1991;10:2669–79.
Ryo A, Liou YC, Lu KP, Wulf G. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J Cell Sci. 2003;116:773–83.
Steger M, Murina O, Hühn D, et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell. 2013;50:333–43.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308.
Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604.
Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J Biol Chem. 2002;277:47976–9.
Desantis A, Bruno T, Catena V, De Nicola F, Goeman F, Iezzi S, et al. Che-1 modulates the decision between cell cycle arrest and apoptosis by its binding to p53. Cell Death Dis. 2015;6:e1764.
Berger M, Stahl N, Del Sal G, Haupt Y. Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol Cell Biol. 2005;25:5380–8.
Sartori AA, Steger M. Prolyl isomerization: a new PIN code for DSB repair. Cell Cycle. 2013;12:2717–8.
Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–8.
Ahmad KA, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2--a key suppressor of apoptosis. Adv Enzym Regul. 2008;48:179–87.
Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta. 2008;1784:33–47.
Homma MK, Homma Y. Cell cycle and activation of CK2. Mol Cell Biochem. 2008;316:49–55.
Messenger MM, Saulnier RB, Gilchrist AD, Diamond P, Gorbsky GJ, Litchfield DW. Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. J Biol Chem. 2002;277:23054–64.
Bandyopadhyay K, Gjerset RA. Protein kinase CK2 is a central regulator of topoisomerase I hyperphosphorylation and camptothecin sensitivity in cancer cell lines. Biochemistry. 2011;50:704–14.
Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22:229–37.
Cheung-Ong K, Giaever G, Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol. 2013;20:648–59.
Faivre S, Chan D, Salinas R, Woynarowska B, Woynarowski JM. DNA strand breaks and apoptosis induced by oxaliplatin in cancer cells. Biochem Pharmacol. 2003;66:225–37.
Suenaga M, Mizunuma N, Matsusaka S, Shinozaki E, Ueno M, Yamaguchi T. Retrospective analysis on the efficacy of bevacizumab with FOLFOX as a first-line treatment in Japanese patients with metastatic colorectal cancer. Asia Pac J Clin Oncol. 2014;10:322–9.
Loupakis F, Cremolini C, Masi G, Lledo G, Flesh M, Mery-Mignard D, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609–18.
Stintzing S, Modest DP, Rossius L, Lerch MM, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol. 2016;17:1426–34.
Polonio-Vallon T, Krüger D, Hofmann TG. ShaPINg cell fate upon DNA damage: role of Pin1 isomerase in DNA damage-induced cell death and repair. Front Oncol. 2014;4:148.
Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–7.
Lu J, Hu Z, Wei S, Lerch MM, von Weikersthal LF, Decker T, et al. A novel functional variant (-842G>C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis. 2009;30:1717–21.
De Oliveira LP, López I, Dos Santos EM, Lerch MM, von Weikersthal LF, Decker T, et al. Association of the p53 codon 72 polymorphism with clinicopathological characteristics of colorectal cancer through mRNA analysis. Oncol Rep. 2014;31:1396–406.
Acknowledgements
The National Institutes of Health (P30CA014089-27S1), the Gloria Borges Wunderglo Project, the Dhont Family Foundation, the Dave Butler Research Fund, and the Call to Cure Research Fund partially supported this work. Mitsukuni Suenaga received a grant from the Takashi Tsuruo Memorial Fund. Martin D. Berger received a grant from the Swiss Cancer League (BIL KLS-3334-02-2014) and the Werner and Hedy Berger-Janser Foundation for Cancer Research. Yuji Miyamoto received a grant from the Japan Society for the Promotion of Science (S2606).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Suenaga, M., Schirripa, M., Cao, S. et al. Potential role of PIN1 genotypes in predicting benefit from oxaliplatin-based and irinotecan-based treatment in patients with metastatic colorectal cancer. Pharmacogenomics J 18, 623–632 (2018). https://doi.org/10.1038/s41397-018-0030-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-018-0030-8