Abstract
Anaerobic oxidation of methane (AOM) reduces methane emissions from marine ecosystems but we know little about AOM in rivers, whose role in the global carbon cycle is increasingly recognized. We measured AOM potentials driven by different electron acceptors, including nitrite, nitrate, sulfate, and ferric iron, and identified microorganisms involved across contrasting riverbeds. AOM activity was confined to the more reduced, sandy riverbeds, whereas no activity was measured in the less reduced, gravel riverbeds where there were few anaerobic methanotrophs. Nitrite-dependent and nitrate-dependent AOM occurred in all sandy riverbeds, with the maximum rates of 61.0 and 20.0 nmol CO2 g−1 (dry sediment) d−1, respectively, while sulfate-dependent and ferric iron-dependent AOM occurred only where methane concentration was highest and the diversity of AOM pathways greatest. Diverse Candidatus Methylomirabilis oxyfera (M. oxyfera)-like bacteria and Candidatus Methanoperedens nitroreducens (M. nitroreducens)-like archaea were detected in the sandy riverbeds (16S rRNA gene abundance of 9.3 × 105 to 1.5 × 107 and 2.1 × 104 to 2.5 × 105 copies g−1 dry sediment, respectively) but no other known anaerobic methanotrophs. Further, we found M. oxyfera-like bacteria and M. nitroreducens-like archaea to be actively involved in nitrite- and nitrate/ferric iron-dependent AOM, respectively. Hence, we demonstrate multiple pathways of AOM in relation to methane, though the activities of M. oxyfera-like bacteria and M. nitroreducens-like archaea are dominant.
Similar content being viewed by others
Introduction
Anaerobic oxidation of methane (AOM) via sulfate reduction reduces methane emissions from marine sediments [1]. Sulfate-dependent AOM is catalyzed by anaerobic methanotrophic archaea of the ANME-1, ANME-2a/2b, ANME-2c, and ANME-3 clades and by sulfate-reducing bacteria. The transfer of reducing equivalents to sulfate is through an interspecies electron carrier or conductive pili (nanowires) [2,3,4], or by excreting zero-valent sulfur compounds for disproportionation by their bacterial partners [5].
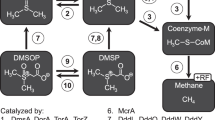
Methane can also be oxidized anaerobically using alternative electron acceptors, e.g., nitrite [6], nitrate [7] and ferric iron [8, 9]. Nitrite-dependent AOM is performed by the NC10 bacteria related to Candidatus Methylomirabilis oxyfera (M. oxyfera) [6, 10], that produces oxygen intracellularly from two nitric oxide molecules for methane oxidation and respiration [10, 11]. A specific lineage of the ANME-2 clade (ANME-2d) (Candidatus Methanoperedens nitroreducens [M. nitroreducens]) is capable of catalyzing AOM through reverse methanogenesis using nitrate [7]. Ettwig et al. [8] showed that M. nitroreducens-like archaea can oxidize methane using ferric iron in addition to nitrate. Very recently, Cai et al. [9] reported a novel genus within the family Candidatus Methanoperedenaceae that can also couple methane oxidation to ferric iron reduction. All these anaerobic methanotrophs have been enriched from various freshwater sediments.
Our knowledge about sulfate-dependent AOM is mainly for marine sediments [12,13,14], as it was assumed to be insignificant in low-sulfate fresh waters (~0.01–0.2 mM versus 28 mM in sea water) [15]. Some have suggested, however, that AOM is coupled to sulfate reduction in lake sediments [16,17,18]. Recently, Weber et al. [19] provided direct evidence that the M. nitroreducens-like archaea are involved in sulfate-dependent AOM in lake sediments through 16S rRNA-based stable isotope probing. Studies of nitrite-dependent AOM in fresh waters are increasing, including wetlands [20, 21], lakes [22,23,24], and reservoirs [25, 26]. Nitrate-dependent AOM could also reduce methane emissions from wetlands [21] and paddy fields [27, 28]. Due to these more diverse electron acceptors in fresh waters, they may support a greater variety of AOM pathways than marine sediments.
Contemporary analyses have increased the estimated contribution from running waters (rivers and streams) to the global methane budget from 1.5 to 26.8 Tg CH4 per year, equivalent to ~15 and 40% of emissions from wetlands and lakes, respectively [29, 30]. Further, our previous work demonstrated that a rise of just 2 °C could increase the proportion of carbon emitted as methane from rivers by 8% [31]. Thus rivers could play an increasing role in the future global methane budget, and our appreciation of rivers as biogeochemical hotspots and major contributors to the global carbon cycle is being revised.
Aerobic methane oxidation reduces methane emissions from rivers [31, 32], but anaerobic oxidation via nitrite, nitrate, sulfate, and ferric iron cannot be excluded. Only molecular evidence for AOM (presence of M. oxyfera-like sequences) has hitherto been reported for rivers [33,34,35], although nitrite-dependent AOM activity was discovered in an enrichment culture from canal sediments [6]. Overall, the potential of different AOM pathways to reduce methane emissions from rivers, the microorganisms involved, are poorly known.
Many of the lake and wetland sediments known to support AOM are probably impermeable silts and clays [20, 21, 23], where diffusion delivers the solutes that sustain microbial metabolism [36]. In contrast, the gravel and sandy sediments which dominate UK riverbeds [37] are more permeable [38, 39], allowing greater advective flux of nutrients (inorganic nitrogen and phosphorus etc.,) and methane through the riverbed. Even though the bulk porewater of such riverbeds has appreciable oxygen, anaerobic metabolism (e.g., anammox and denitrification) can occur in anoxic microsites [39, 40]. The physical heterogeneity of the riverbeds and the co-occurrence of methane and nitrate reduction, along with other electron acceptors [39, 41], could allow the co-existence of various AOM communities. Further, the contrasting characteristics of gravels and sands (e.g., the porewater methane concentration in sands is far greater than that in gravels [39, 42]) in turn, could affect the distribution and activity of AOM communities.
Our aim was to characterize the significance of AOM pathways driven by the electron acceptors nitrite, nitrate, sulfate, and ferric iron in reducing methane emissions, and to identify the anaerobic methanotrophs actively involved along a gradient of declining methane concentration from sandy to gravel riverbeds.
Materials and methods
Sample collection
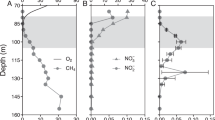
Sediment and porewater were collected from seven riverbeds in southeast England between February 2016 and January 2017 (Table S1). The rivers Lambourn and Stour (I and II) have predominantly gravel beds (grain size 2–16 mm) and overlie permeable chalk geology, while the Hammer Stream and rivers Medway, Marden, and Nadder have predominantly sandy beds (grain size 0.062–2 mm) which are typically less permeable [43, 44]. In each river, six intact sediment cores were collected to a depth of 16 cm and porewaters (five replicates) sampled using mini-probes [38]. Porewater samples for NH4+, NO2−, and NO3− analyses were preserved by filtering (0.2 μm polypropylene) and samples for methane (CH4) transferred to vials (3 ml, Labco, UK) and preserved with ZnCl2 (100 μl, 50% (w/v)). Surface waters were treated in the same way and all samples were kept in cool bags (Thermos) for transfer to the laboratory (<4 h). Sediment cores were then sliced in an anoxic glove box (AGB) (O2 < 100ppm; Belle Technology, Dorset, UK) at five depths horizons: 0–2, 2, 4–8, 8–12, and 12–16 cm. This study is supported in part by published methane and Fe2+ data (Fig. 1a, b) generated by a study of in situ nitrogen cycling in the Hammer Stream [41] and a wider study of the biogeochemistry in rivers on chalk, sand, and clay geologies [39, 43, 45].
Porewater a methane (ln, natural logarithm) and b Fe2+ concentrations across contrasting sandy and gravel riverbeds (data from ref. [41]; and unpublished from ref. [39], which are available at 10.5285/7ded510f-3955- 4b92-851d-29c0f79a0b99) and c PCA ordination diagram of porewater chemisty within our present riverbeds
Physicochemical analyses
Surface waters and porewaters were transferred through a three-way stopcock into a syringe holding a pH or O2 probe [38] and the concentrations of NH4+, NO2−, and NO3− measured using an auto-analyzer (Skalar San+2, Breda, The Netherlands). Methane concentration was measured by gas chromatography (GC/FID), as described previously [31].
Isotope tracer experiments
AOM activity was measured using 13CH4. Approximately 2−3 g of sediments and 5 ml of N2-degassed synthetic river water (0.12 g l−1 NaHCO3, 0.04 g l−1 KHCO3, 0.027 g l−1 MgCl2, and 0.092 g l−1 CaCl2·2H2O) were transferred into 12 ml vials (Labco, UK) inside the AGB and then preincubated for 2d on orbital shakers to remove background NOx− (NO2− + NO3−). After preincubation, the vials were injected with 100 μl of N2-degassed stock solutions of either NaNO2 (1 mM), NaNO3 (2.5 mM), FeCl3 (1 mM) or Na2SO4 (1 mM). Finally, 50 μl of 13CH4 was injected to give a ~1% (v/v) of methane headspace and control slurries were left unamended. Our incubations to measure AOM potential lasted less than 4 weeks, during which six replicate vials were sacrificed (300 μl, ZnCl2, 50% (w/v)) at different intervals and headspace concentrations of 12CO2 and 13CO2 (m/z 44 and 45) measured using CF/IRMS. The overlying water in each vial was then collected and acidified (HCl, 100 μl, 12.2 M), and DIC (CO2 + HCO3− + CO32−) measured as 12CO2 or 13CO2 [32], where 13CO2 plus 13C-DIC equalled the total amount of 13C-CH4 oxidized to inorganic carbon. An obvious lag phase was observed before 13CO2 production in slurries amended with 13CH4 + NO2− or NO3− (Results section), therefore AOM activity was calculated by linear regression after the lag phase. Having demonstrated a potential for AOM, we then repeated the incubations using a shorter preincubation period (6 h) to see if we could remove any lag phase and measure the potentials closer to in situ (Results section).
To determine organic 13C content, the incubated sediments were acidified (as above) to remove DIC, dried to a constant mass (60 °C) and then combusted at 1000 °C in an integrated elemental analyzer and mass spectrometer (Sercon Integra 2). The AOM carbon conversion efficiency (CCE) was calculated as the fraction of total 13C-CH4 oxidized to organic carbon.
DNA extraction
Genomic sediment DNA was extracted using a Power Soil DNA Isolation Kit (MO-BIO-Laboratories, USA) and its concentration and quality measured with a NanoDrop (ND-1000; Isogen Life Science, the Netherlands).
PCR amplification and Illumina Miseq sequencing
The hypervariable regions of the bacterial (V3–V4) and archaeal (V4–V5) 16S rRNA genes were amplified with the universal primer sets 319f-806r [46] and 524f-958r [47], respectively. The preparation of the PCR mixtures and thermal programmes for the bacteria and archaea were as described previously [21, 48]. Total bacterial and archaeal PCR products were gel purified using QIAquick gel extraction kits (Qiagen, Chatsworth, California, USA) and sequenced using the 300 bp paired-end strategy on the Illumina MiSeq platform. The downstream sequence analyses were conducted using the Quantitative Insights into Microbial Ecology (QIIME) [49]. The reads from the original DNA fragments were merged, and reads with an average quality score above 25 were accepted. The occurrence of chimeric sequences was further examined using QIIME. High-quality reads were then classified with the Ribosomal Database Project (RDP) classifier using the SILVA databases.
Phylogenetic analysis
Phylogenetic analysis of the sequences was performed using Mega 5 software; evolutionary distance computed using the Maximum Likelihood and tree topology tested using bootstrap analysis (1000 replicates).
Quantitative PCR
The 16S rRNA gene abundance of total bacteria and total archaea was determined using the primer sets 341f-518r [50] and Arch967f-Arch1060r [51], respectively. Primer sets of qp1f-qp1r [52] and 641F-834R [17] were used for quantification of the 16S rRNA genes of M. oxyfera-like bacteria and M. nitroreducens-like archaea, respectively (see Table S1 for details of quantitative PCR (qPCR) primers). The qPCR was performed on an iCycler iQ5 thermocycler (BioRad) with standard curves constructed from a serial dilution of a known copy number of the plasmid DNA. Triplicate qPCRs analyses were performed for each sample and each dilution and the specialties of qPCR products examined by melt curve analysis with a detection limit of 9.2–10.8 copies per well.
Real-time quantitative reverse transcription PCR
Total RNA was extracted using a RNA PowerSoil Total RNA Isolation Kit (Qiagen, USA) and treated with a DNA-free™ DNA Removal Kit (Invitrogen, USA). RNA concentration was quantified with a Qubit 2.0 Fluorometer (Invitrogen, USA) and total RNA then used for cDNA synthesis using Super-ScriptTM II reverse transcriptase (Invitrogen, USA). Transcript abundance of M. oxyfera-like bacterial pmoA mRNA and M. nitroreducens-like archaeal mcrA mRNA were quantified in conjunction with the tracer experiments using the primer sets cmo182-cmo568 [53] and McrA159f-McrA345r [35], respectively. The RT-qPCR was conducted as for qPCR described above using BioRad CFX 384, with a detection limit of 9.8–10.7 copies per well.
Results
Porewater chemistry
Porewater methane in sandy riverbeds (0.7–354.3 μM) was significantly higher (t-test, p < 0.05) than that in the gravels (0.02–34.0 μM), and tended to increase with depth (Fig. S1). In contrast, oxygen was significantly lower (t-test, p < 0.05) in the sand (38.7–133.3 μM) compared to the gravel (92.6–288.7 μM) (Fig. S1) and, overall, the sandy Hammer Stream had the highest methane with the lowest oxygen. Porewater nitrite was 0.5–5.9 μM across the riverbeds, and was highest in the upper 2 cm sediments (Fig. S1). Nitrate concentration decreased with depth in the sand (8–116 μM) but increased with depth in the gravel (57–489 μM), reflecting the stronger influence of groundwater on the more permeable chalk. Ammonium concentration was low in the Lambourn (0.7–1.2 μM) but extremely high in the Medway (476.0–2354.4 μM) (Fig. S1). Elsewhere ammonium concentration was 7.9–184.5 μM. Shelley et al. [41] reported mean Fe2+ concentrations in the Hammer Stream sediments of 178 μM, and data generated by Lansdown et al. [39] clearly show higher concentrations for Fe2+ in sandy riverbeds (136.9 μM) than in gravels (17.0 μM) (Fig. 1b). According to the present and previously reported data (Table 1), we define sandy riverbeds as being more reduced than gravels, with on average about 10 times more methane, 8 times more Fe2+, and 50% less oxygen. Further, principle component analysis (PCA) of our present riverbeds showed sharp separation of oxygen and methane on PC1 (accounted for 59% of the variance, Fig. 1c), suggesting that this axis represented a chemical gradient moving from more reduced sandy riverbeds to less reduced gravel riverbeds.
AOM potential driven by different electron acceptors
The 13CH4 tracer experiments showed no significant production of 13CO2 in any of the gravel sediments amended with any electron acceptor (NO2−, NO3−, SO42−, Fe3+) (Fig. S2). In contrast, both NO2− and NO3− generated 13CO2 with all sandy sediments; though there was an obvious lag phase before 13CO2 production began (Fig. 2). Different ranges of nitrite-dependent AOM activity were observed (Fig. 3): the potential in the Hammer Stream (12.2–61.0 nmol CO2 g−1 [dry sediment] d−1) was significantly higher (t-test, p < 0.05) than that measured in the Medway, Marden, and Nadder, with activities of 1.4–10.9, 1.2–7.0, and 0.4–1.1 nmol CO2 g−1 (dry sediment) d−1, respectively. Nitrate-dependent AOM activity was lower than that of nitrite-dependent AOM, except for the River Nadder (Fig. 3). Nitrate-dependent AOM activities were 5.4–20.0, 0.5–4.3, 0.6–1.7, and 0.8–5.7 nmol CO2 g−1 (dry sediment) d−1, respectively, in the Hammer Stream and rivers Medway, Marden, and Nadder. Having shown the AOM potentials driven by nitrite and nitrate with a lag phase, we collected more sediment from two representative rivers (Hammer Stream and River Nadder) for a second set of experiments with a shorter, 6 h preincubation. Here, there was no lag phase for NOx−-dependent AOM and the activity was similar to that after a 2d preincubation (Fig. 2).
Examples of 13CO2 production in slurries of sediment from the 4 to 8 cm depth horizon amended with 13CH4 and different electron acceptors after both short and long preincubations. a and c both durations in the Hammer stream and River Nadder, respectively, and b and d long preincubations only for the rivers Medway and Marden, respectively
The addition of both SO42−and Fe3+ stimulated 13CO2 production in the Hammer Stream (Fig. 2; Table 1). Sulfate-dependent and ferric iron-dependent AOM activities were 0.6–4.4 and 1.5–8.1 nmol CO2 g−1 (dry sediment) d−1, respectively, at different depths in the riverbed.
Organic 13C could only be measured in slurries amended with 13CH4 + NO2− in sediment from the Hammer Stream, where the CCE was 8.2% on average. Therefore, although all the sandy riverbeds exhibited an AOM potential, only the most reduced Hammer Stream provided evidence for a growing AOM community.
Expression analyses of pmoA mRNA and mcrA mRNA
After 4 weeks of incubation we assayed sediments from the Hammer Stream (4–8 cm layer), both with and without additions of 13CH4, 13CH4 + NO2−, 13CH4 + NO3−, 13CH4 + SO42−, and 13CH4 + Fe3+, for gene expression. Similarly, sediments from the River Nadder (2−4 cm layer) were also screened for expression but only in the incubations with 13CH4 + NO2− and 13CH4 + NO3−, as only they had produced measurable amounts of 13CO2.
The abundances of pmoA transcripts in slurries amended with 13CH4 + NO2− (8.7 × 104 copies g−1 dry sediment) and 13CH4 + NO3−(4.9 × 104 copies g−1 dry sediment) were higher (t-test, p < 0.05) than those amended with 13CH4, 13CH4 + SO42−, 13CH4 + Fe3+, and controls (4.6–7.0 × 103 copies g−1 dry sediment) in the Hammer Stream (Fig. 4a). Similarly, in the River Nadder (Fig. 4a), higher abundances (t-test, p < 0.05) of pmoA transcripts were observed in slurries amended with 13CH4 + NO2− (2.4 × 104 copies g−1 dry sediment) and 13CH4 + NO3−(1.9 × 104 copies g−1 dry sediment), than in those amended with only 13CH4 and the controls (4.2–5.4 × 103 copies g−1 dry sediment).
In the Hammer Stream (Fig. 4b), the abundances of M. nitroreducens-like archaeal mcrA transcripts were higher (t-test, p < 0.05) in slurries amended with 13CH4 + NO3− and 13CH4 + Fe3+ (6.0 and 8.7 × 103 copies g−1 dry sediment, respectively) than in the other treatments or controls (1.2–1.9 × 103 copies g−1 dry sediment). In the River Nadder, higher (t-test, p < 0.05) transcript abundance of mcrA was also detected, but only in slurries amended with 13CH4 + NO3− (7.1 × 103 copies g−1 dry sediment), compared to the other treatments and controls (1.7–2.1 × 103 copies g−1 dry sediment).
Further, the pmoA cDNA derived from slurries amended with 13CH4 + NO2− and 13CH4 + NO3−, and for mcrA in slurries amended with 13CH4 + NO3− and 13CH4 + Fe3+, was cloned and sequenced. Phylogenetic analysis showed that the pmoA cDNA (26 sequences) formed a distinct cluster (>99% identity between each other) in the tree, showing distant identity (89.4–90.5%) to the pmoA gene of Candidatus Methylomirabilis oxyfera [10] but a higher identity (97–99%) to the pmoA genes reported for reservoir sediments (Fig. S3). Three different clusters of the mcrA cDNA (18 sequences) were present in the phylogenetic tree (Fig. S4), showing 92.5–95.2 and 93.0–98.9% identity to the mcrA genes of Candidatus Methanoperedens nitroreducens (JMIY0100002) [7] and Candidatus Methanoperedens sp. BLZ1 [54], respectively. The three clusters were also closely related (>95% identity) to the mcrA genes reported in paddy soils [27] and river sediment [35].
Bacterial and archaeal communities across the riverbeds
The microbial community composition at different depths of the sandy riverbeds, in which AOM activity was detected, was examined by Illumina sequencing of the total bacterial and archaeal 16S rRNA genes. After tag merge, removal of chimeric sequences and quality control, the ranges of total (clean) bacterial and archaeal sequences were 42,505–64,875 and 30,287–42,758, respectively (Table 2).
Proteobacteria was the most abundant phylum across all riverbeds (Fig. S5). The percentage of M. oxyfera-like sequences in the Hammer Stream (0.8–2.1%) was higher than that in other riverbeds (0.3–0.9%; Table 2). The taxonomic affiliations of the sequences for Type I and Type II aerobic methanotrophs were compared at the order, family and genus levels [55, 56] which showed that they comprised <0.1% of the total reads. No sequences related to Verrucomicrobia methanotrophs (Methylacidiphilum) [57] could be detected.
The phylum Bathyarchaeota was the dominant archaea in the Hammer Stream and the rivers Medway and Nadder (Fig. S6). In the River Marden, Thaumarchaeota was the dominant phylum. A relatively higher occurrence of M. nitroreducens-like sequences was found in the Hammer Stream and River Nadder, accounting for 1.5–2.7 and 2.2–6.5%, respectively, of the total archaeal sequences in each sample (Table 2). No members of the marine anaerobic methanotrophs (ANME-1, ANME-2a/2b, ANME-2c, and ANME-3) were detected.
Phylogenetic analyses of known anaerobic methanotrophs
Phylogenetic analysis showed that the M. oxyfera-like sequences recovered were 90–97% identical to the 16S rRNA gene of Candidatus Methylomirabilis oxyfera [10]. These sequences were closely related (>96% identical) to the M. oxyfera-like sequences reported in enrichment cultures [52], lake sediments [23, 58, 59], and paddy soil [27] (Fig. 5).
Phylogenetic analysis of M. nitroreducens-like sequences showed that they were closely related (>96% identical) to the M. nitroreducens-like sequences from enrichment cultures [60, 61], lake sediments [19], and paddy soils [27] (Fig. 6).
Maximum likelihood phylogenetic tree showing the affiliations of the 16S rRNA genes of M. nitroreducens-like archaea (449 bp) recovered from the sandy riverbeds. Bootstrap values > 50% (out of 1000 replicates) are shown in front of respective nodes, and the scale bar represents 2% sequence divergence
Quantification of known anaerobic methanotrophs
The 16S rRNA gene abundance of total bacteria (1.8 × 108 to 1.1 × 109 copies g−1 dry sediment) was higher (t-test, p < 0.05) than that of archaea (4.0 × 106 to 2.9 × 107 copies g−1 dry sediment) in all of the riverbeds (Fig. 7). The highest 16S rRNA gene abundance of M. oxyfera-like bacteria was detected in the Hammer Stream (3.7 × 106 to 1.5 × 107 copies g−1 dry sediment). Very low M. oxyfera-like bacterial 16S rRNA gene abundance was measured in the gravel riverbeds (<8.7 × 103 copies g−1 dry sediment). The 16S rRNA gene abundance of M. oxyfera-like bacteria correlated well with the potential nitrite-dependent AOM rates (Fig. S7). The ratios of gene abundance of M. oxyfera-like bacteria to total bacteria and total bacteria plus total archaea were 0.5–2.8 and 0.1–2.7%, respectively, in the sands.
The 16S rRNA gene abundance of M. nitroreducens-like archaea was 2.1 × 104 to 2.5 × 105 copies g−1 dry sediment in the sands, while its abundance in the gravels was extremely low (<2.5 × 103 copies g−1 dry sediment). Among these riverbeds, the River Nadder contained a relatively greater abundance of M. nitroreducens-like archaeal 16S rRNA genes (Fig. 7). The ratios of gene abundance of M. nitroreducens-like archaea to total archaea and total bacteria plus total archaea were 0.09–1.6 and 0.003–0.07%, respectively, in the sands.
Discussion
The potential for several pathways of AOM was detected in sandy riverbeds. Nitrite and nitrate are the dominant electron acceptors for AOM, although sulfate and ferric iron both stimulated AOM in the Hammer Stream where methane concentration was highest (Table 1). Further, we provide evidence for the active involvement of M. oxyfera-like bacteria and M. nitroreducens-like archaea in nitrite- and nitrate/ferric iron-dependent AOM, respectively, in sandy riverbeds.
The significance of AOM in rivers is largely unexplored. For example, only molecular evidence of M. oxyfera-like bacteria has been reported previously, with no tracer measurements of activity [33,34,35]. Here, AOM activity was prevalent in the more reduced sandy riverbeds, whereas no activity could be detected in the more oxygenated gravel riverbeds. Both NO2− and NO3− stimulated AOM in all sandy riverbeds, and stimulation by NO2− was greater than by NO3− (Fig. 2). No obvious lag phase was observed for either nitrite- or nitrate-dependent AOM during the tracer experiments with a shorter preincubation time (6 h) and the activity (1.2–53.6 nmol CO2 g−1 [dry sediment] d−1) was similar to that (0.4–61.0 nmol CO2 g−1 [dry sediment] d−1) measured in experiments with longer preincubation times (2d) (Fig. 2), and the activity probably represented in situ potential. The rate of nitrite-dependent AOM measured in our rivers, particularly in the Hammer Stream, is considerably greater than that reported in lake sediments (1.8–3.6 nmol CO2 ml−1 sediment d−1) [23] (activity was determined after 3–4 weeks of incubation), or in wetland sediments (~10 nmol CO2 g−1 [dry soil] d−1; activity was determined within 1 day of incubation) [20, 21]. Zhu et al. [62] reported a potential nitrite-dependent AOM rate of 9 nmol CO2 g−1 (dry soil) d−1 in peatland after 3 months of incubation, with no activity being detected within the first 2 weeks. Furthermore, Deutzmann et al. [24] reported that the potential nitrite-dependent AOM rate, which was calculated from micro-sensor profiles, can be up to 40 nmol CH4 cm−3 d−1 in a lake water column. Here, a low porewater nitrite concentration was detected in our riverbeds (Fig. S1), which may be due to it being rapidly reduced either through AOM or anammox as, at times, nitrite is hard to detect at all [39]. The Hammer Stream sediments are known to support high in situ rates of nitrate reduction, with the majority being due to denitrification [41] and denitrification also dominates in the rivers Nadder and Marden [39]. Therefore, the nitrite required for nitrite-dependent AOM in the sandy riverbeds may mainly derive from denitrification.
In lake and wetland sediments in which activity of nitrite-dependent AOM has also been reported, the sediments were probably impermeable silts and clays [20, 21, 23] where the transport of solutes is dominated by diffusion. In contrast, our gravel and sandy riverbeds are more permeable, allowing a greater exchange of solutes (NOx−) and dissolved methane [36] that could sustain a relatively higher potential of nitrite-dependent AOM. Although under maximum permeability in the gravels, the fast delivery of oxygen and low availability of methane appears unsuitable for the persistence of an active nitrite-dependent AOM community. In addition, 13C-carbon assimilation from 13CH4 was only measurable in the Hammer Stream; although we cannot exclude assimilation of any 13CO2 by other autotrophic organisms besides that assimilated directly by the M. oxyfera-like bacteria. Here, the CCE (8.2%) is lower than that previously reported for aerobic methanotrophs (~50%) in gravel riverbeds [32] but consistent with a CCE of 9% for an enrichment culture of Candidatus Methylomirabilis oxyfera [63], suggesting that the most reduced sediments of the Hammer Stream facilitate nitrite-dependent AOM.
Our nitrate-dependent AOM activity ranged between 0.5 and 20.0 nmol CO2 g−1 (dry sediment) d−1, lower than the nitrite-dependent AOM activity. This nitrate-dependent AOM activity is higher than that measured previously (within 24 h of incubation) in freshwater marsh sediments (0.4–1.2 nmol CO2 g−1 [dry sediment] d−1 [21]), but lower than that for paddy soils (e.g., 79.9 nmol CO2 g−1 [dry soil] d−1 [27]) where the activity was determined after 4 weeks of incubation following an obvious lag phase. It should be noted, however, that methane oxidation is not directly coupled only to NO3− reduction in slurries amended with 13CH4 + NO3− because both mcrA and pmoA genes were highly expressed in these slurries (Fig. 4) and some NO3− may first be partially reduced to NO2− (e.g., denitrification) and the NO2− then used for methane oxidation.
We could measure comparatively little methane in the surface waters of our gravel (0.05–0.3 μM) and sandy rivers (0.3–5.1 μM) (Table S3), whereas methane was up to 100 times more concentrated in the sediment porewaters (~34.0 and ~354 μM, respectively, for the gravels and sands). Further, with more porewater methane data for the Hammer Stream and River Nadder (Fig. 1a), it is evident that the exponential decay in methane of 83–95% over the top 5 to 25 cm of sediment coincides with the in situ peak in nitrate reduction [41]. We have also measured aerobic methane oxidation in the sandy rivers here (except the Medway; Fig. S8) and 32 gravel rivers previously [64], with similar activity (1.7–47.4 nmol CH4 g−1 [dry sediment] d−1) to that of nitrite-dependent AOM. However, given that nitrate is typically more abundant than nitrite [65], it is more reasonable to use the AOM potentials measured with 13CH4 + NO3− to estimate the role of NOx−-dependent AOM in attenuating methane emissions from sandy riverbeds. Accordingly, for the sandy riverbeds that are common in the UK (~26% of 9459 sites [37]) and probably beyond, we estimate that microbial methane oxidation is about 35% anaerobic and 65% aerobic. In more permeable and widespread gravel beds (48% of UK sites; [37]), however, methane oxidation is carried out almost exclusively by aerobic methanotrophs [32, 64, 66].
In addition to NO2− and NO3−, SO42−, and Fe3+ also stimulated AOM in the most reduced sediment from the Hammer Stream (Table 1), suggesting that high methane concentrations support a greater diversity of AOM pathways; although the potential rates for SO42− and Fe3+-dependent AOM were lower than NOx−-dependent AOM. The standard Gibbs free energies for nitrite- (−928 kJ mol−1) and nitrate-dependent AOM (−519.8 kJ mol−1) are greater than those for sulfate- (−16.6 kJ mol−1) and ferric iron-dependent AOM (−81.6 kJ mol−1) [67] and NOx−-dependent AOM is energetically more favorable. Thus, NOx−-dependent methanotrophs may have an advantage in using methane over the sulfate- and ferric iron-dependent methanotrophs.
Any methane still present below the dominant zone of nitrate reduction could also be used by sulfate- and ferric iron-dependent anaerobic methanotrophs, as here in the Hammer Stream. The Hammer Stream sediments are rich in Fe2+ (~178 µM) [41], which could be primarily from Fe3+ reduction, further suggesting that the Hammer Stream is a good environment for ferric iron-dependent AOM. The activity of sulfate- and ferric iron-dependent AOM (0.6–4.4 and 1.5–8.1 nmol CO2 g−1 [dry sediment] d−1, respectively) was at the lower end (3–36 nmol CO2 g−1 [dry sediment] d−1) of that reported in lake sediments, where AOM is probably coupled to sulfate and/or ferric iron reduction and NOx− is comparatively scarce [16, 19].
To identify the AOM-mediating methanotrophs, the transcript abundance of M. oxyfera-like bacterial pmoA and M. nitroreducens-like archaeal mcrA was analyzed in conjunction with the 13C-tracer incubations. Our approach is therefore different from the most recent reports, in which AOM activity in lake and wetland sediments was linked to M. oxyfera-like bacteria or M. nitroreducens-like archaea only by detecting the presence of their specific 16S rRNA and/or functional genes [20, 21, 23, 24, 27]. The linear production of 13CO2 throughout the second set of tracer experiments, and prior to the gene expression experiments (Fig. 2), suggested that the microbial community did not change significantly during our incubations. Here, the addition of 13CH4 and NO2−or NO3− significantly stimulated the expression of M. oxyfera-like bacterial pmoA (Fig. 4a), strongly suggesting that M. oxyfera-like bacteria are actively involved in nitrite-dependent AOM in our sandy riverbeds. The presence of pmoA genes with <93% nucleic acid sequence identity, compared to known species, has been taken to indicate novel methanotrophs [68]. Our recovered pmoA formed a distinct cluster only distantly related (89.4–90.5%) to the pmoA gene of Candidatus Methylomirabilis oxyfera [10], indicating that a potentially novel M. oxyfera cluster (or perhaps a novel M. oxyfera strain/species) is reducing methane emissions from sandy riverbeds. In addition to the M. oxyfera-like bacteria, some aerobic methanotrophs can use NO2−/NO3− as alternative electron acceptors under oxygen limitation [69, 70]. Such aerobic methanotrophs may also play a role in methane oxidation here.
A significantly higher transcript abundance of M. nitroreducens-like archaeal mcrA was detected in slurries amended with 13CH4 + NO3− (Fig. 4b), suggesting that these archaea are active and responsible for nitrate-dependent AOM in these sandy sediments. Also, the addition of Fe3+ stimulated the expression of M. nitroreducens-like archaeal mcrA (Fig. 4b) and, therefore, these archaea are probably responsible for ferric iron-coupled AOM in the Hammer Stream. Clusters I and II of the mcrA were both detected in the sediment incubations from the Hammer Stream and River Nadder amended with 13CH4 + NO3−, while cluster III could only be detected by addition of 13CH4 + Fe3+ to sediments from the Hammer Stream (Fig. S4). This indicates that clusters I and II were probably responsible for nitrate-dependent AOM, while cluster III was probably responsible for ferric iron-dependent AOM. Unlike M. oxyfera-like bacteria, which can only use nitrite, M. nitroreducens-like archaea may be more flexible, with the potential to use several electron acceptors.
No members of the marine anaerobic methanotrophs were detected in any of our samples and are unlikely to be major participants in sulfate-dependent AOM; though their abundance may have been too low to detect. Recent evidence suggests that M. nitroreducens-like archaea are involved in sulfate-dependent AOM in lake sediments [19] but here the addition of sulfate did not stimulate the expression of mcrA of M. nitroreducens-like archaea (Fig. 4b). However, we cannot exclude M. nitroreducens-like archaea performing sulfate-dependent AOM in our sandy riverbeds, because the specific mcrA primers used here may not have covered all lineages. Some 16S rRNA gene sequences of M. nitroreducens-like archaea recovered from the riverbeds were closely related (96–99% identical) to sequences reported from Lake Ørn (Fig. 6), where M. nitroreducens-like archaea are involved in sulfate-dependent AOM [19].
The 16S rRNA gene abundance of M. oxyfera-like bacteria (9.3 × 105 to 1.5 × 107 copies g−1 dry sediment) was similar to that previously reported from river (106 to 107 copies g−1 sediment) [34] and lake sediments (105 to 106 copies g−1 sediment) [59]. Here, the 16S rRNA gene abundance of M. nitroreducens-like archaea (2.1 × 104 to 2.5 × 105 copies g−1 dry sediment) was lower (t-test, p < 0.05) than that of M. oxyfera-like bacteria, which agrees with the lower nitrate-dependent AOM potentials, compared to nitrite-dependent AOM. The 16S rRNA gene abundance of M. nitroreducens-like archaea is higher than that in freshwater marsh sediments (103 to 104 copies g−1 dry sediment) [21], but lower than that in paddy soils (105 to 106 copies g−1 dry soil) [27].
In summary, we provide the first evidence of multiple active pathways of AOM driven by different electron acceptors in reduced riverbeds, including nitrite, nitrate, sulfate, and ferric iron. Among these, nitrite and nitrate-dependent AOM could be the most important in regulating methane emissions from more reduced sandy riverbeds, relative to more permeable and more oxygenated gravel beds. The diversity of AOM pathways is greatest where methane concentration is highest, suggesting a link between the diversity of AOM pathways and the availability of methane.
References
Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–34.
McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 2015;526:531–5.
Meulepas RJW, Jagersma CG, Khadem AF, Stams AJM, Lens PNL. Effect of methanogenic substrates on anaerobic oxidation of methane and sulfate reduction by an anaerobic methanotrophic enrichment. Appl Microbiol Biotechnol. 2010;87:1499–506.
Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature. 2015;526:587–90.
Milucka J, Ferdelman TG, Polerecky L, Franzke D, Wegener G, Schmid M, et al. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature. 2012;491:541–6.
Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–21.
Haroon M, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–70.
Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MS, Kartal B. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci USA. 2016;113:12792–6.
Cai C, Leu AO, Xie GJ, Guo J, Feng Y, Zhao JX, et al. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 2018;12:1929–39.
Ettwig K, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8.
Wu ML, de Vries S, van Alen TA, Butler MK, Op den Camp HJ, Keltjens JT, et al. Physiological role of the respiratory quinol oxidase in the anaerobic nitrite-reducing methanotroph ‘Candidatus Methylomirabilis oxyfera’. Microbiology. 2011;157:890–8.
Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–6.
Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci USA. 2002;99:7663–8.
Schubert CJ, Coolen MJ, Neretin LN, Schippers A, Abbas B, Durisch-Kaiser E, et al. Aerobic and anaerobic methanotrophs in the Black Sea water column. Environ Microbiol. 2006;8:1844–56.
Canfield DE, Farquhar J. Animal evolution, bioturbation, and the sulfate concentration of the oceans. Proc Natl Acad Sci USA. 2009;106:8123–7.
Norði Kà, Thamdrup B, Schubert CJ. Anaerobic oxidation of methane in an iron-rich Danish freshwater lake sediment. Limnol Oceanogr. 2013;58:546–54.
Schubert CJ, Vazquez F, Losekann-Behrens T, Knittel K, Tonolla M, Boetius A. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol Ecol. 2011;76:26–38.
Weber HS, Thamdrup B, Habicht KS. High sulfur isotope fractionation associated with anaerobic oxidation of methane in a low-sulfate, iron-rich environment. Front Earth Sci. https://doi.org/10.3389/feart.2016.00061. 2016.
Weber HS, Habicht KS, Thamdrup B. Anaerobic methanotrophic archaea of the ANME-2d cluster are active in a low-sulfate, iron-rich freshwater sediment. Front Microbiol. 2017;8:619 https://doi.org/10.3389/fmicb.2017.00619
Hu B, Shen LD, Lian X, Zhu Q, Liu S, Huang Q, et al. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci USA. 2014;111:4495–500.
Shen LD, Wu HS, Liu X, Li J. Cooccurrence and potential role of nitrite- and nitrate-dependent methanotrophs in freshwater marsh sediments. Water Res. 2017;123:162–72.
Graf JS, Mayr MJ, Marchant HK, Tienken D, Hach PF, Brand A et al. Bloom of a denitrifying methanotroph, “Candidatus Methylomirabilis limnetica”, in a deep stratified lake. Environ Microbiol. https://doi.org/10.1111/1462-2920.14285. 2018.
Deutzmann JS, Schink B. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl Environ Microbiol. 2011;77:4429–6.
Deutzmann JS, Stief P, Brandes J, Schink B. Anaerobic methane oxidation coupled to denitrification is the dominant methane sink in a deep lake. Proc Natl Acad Sci USA. 2014;111:18273–8.
Kojima H, Tokizawa R, Kogure K, Kobayashi Y, Itoh M, Shiah FK, et al. Community structure of planktonic methane-oxidizing bacteria in a subtropical reservoir characterized by dominance of phylotype closely related to nitrite reducer. Sci Rep. 2014;4:5728 https://doi.org/10.1038/srep05728
Naqvi SWA, Lam P, Narvenkar G, Sarkar A, Naik H, Pratihary A, et al. Methane stimulates massive nitrogen loss from freshwater reservoirs in India. Nat Commun. 2018;9:1265.
Vaksmaa A, Luke C, van Alen T, Vale G, Lupotto E, Jetten MS, et al. Distribution and activity of the anaerobic methanotrophic community in a nitrogen-fertilized Italian paddy soil. FEMS Microbiol Ecol. 2016;92:pii: fiw181.
Vaksmaa A, van Alen TA, Ettwig KF, Lupotto E, Valè G, Jetten MS, Lüke C. Stratification of diversity and activity of methanogenic and methanotrophic microorganisms in a nitrogen-fertilized Italian paddy soil. Front Microbiol. 2017;8:2127 https://doi.org/10.3389/fmicb.2017.02127
Bastviken D, Tranvik LJ, Downing JA, Crill PM, Enrich-Prast A. Freshwater methane emissions offset the continental carbon sink. Science. 2011;331:50.
Stanley EH, Casson NJ, Christel ST, Crawford JT, Loken LC, Oliver SK. The ecology of methane in streams and rivers: patterns, controls, and global significance. Ecol Monogr. 2016;86:146–71.
Shelley F, Abdullahi F, Grey J, Trimmer M. Microbial methane cycling in the bed of a chalk river: oxidation has the potential to match methanogenesis enhanced by warming. Freshwat Biol. 2015;60:150–60.
Trimmer M, Shelley FC, Purdy KJ, Maanoja ST, Chronopoulou P-M, Grey J. Riverbed methanotrophy sustained by high carbon conversion efficiency. ISME J. 2015;9:2304–14.
Long Y, Jiang X, Guo Q, Li B, Xie S. Sediment nitrite-dependent methane-oxidizing microorganisms temporally and spatially shift in the Dongjiang River. Appl Microbiol Biotechnol. 2017;101:401–10.
Shen L, Liu S, Zhu Q, Li XY, Cai C, Cheng DQ, et al. Distribution and diversity of nitrite-dependent anaerobic methane-oxidising bacteria in the sediments of the Qiantang River. Microb Ecol. 2014;67:341–9.
Vaksmaa A, Jetten MSM, Ettwig KF, Lüke C. McrA primers for the detection and quantification of the anaerobic archaeal methanotroph ‘Candidatus Methanoperedens nitroreducens’. Appl Microbiol Biotechnol. 2017;101:1631–41.
Huettel M, Rusch A. Transport and degradation of phytoplankton in permeable sediment. Limnol Oceanogr. 2000;45:534–49.
Naura M, Clark MJ, Sear DA, Atkinson PM, Hornby DD, Kemp P, et al. Mapping habitat indices across river networks using spatial statistical modelling of River Habitat Survey data. Ecol Indic. 2016;66:20–29.
Lansdown K, Heppell CM, Dossena M, Ullah S, Heathwaite AL, Binley A, et al. Fine-scale in situ measurement of riverbed nitrate production and consumption in an armored permeable riverbed. Environ Sci Technol. 2014;48:4425–34.
Lansdown K, McKew BA, Whitby C, Heppell CM, Dumbrell AJ, Binley A, et al. Importance and controls of anaerobic ammonium oxidation influenced by riverbed geology. Nat Geosci. 2016;9:357–60.
Stelzer RS, Bartsch LA, Richardson WB, Strauss EA. The dark side of the hyporheic zone: depth profiles of nitrogen and its processing in stream sediments. Freshwat Biol. 2011;56:2011–33.
Shelley F, Klaar M, Krause S, Trimmer M. Enhanced hyporheic exchange flow around woody debris does not increase nitrate reduction in a sandy streambed. Biogeochemistry. 2017;136:353–72.
Trimmer M, Hildrew AG, Jackson MC, Pretty JL, Grey J. Evidence for the role of methane-derived carbon in a free-flowing, lowland river food web. Limnol Oceanogr. 2009;54:1541–7.
Heppell CM, Binley A, Trimmer M, Darch T, Jones A, Malone E, et al. Hydrological controls on DOC: nitrate resource stoichiometry in a lowland, agricultural catchment, southern UK. Hydrol Earth Syst Sci. 2017;21:4785–802.
Pretty JL, Hildrew AG, Trimmer M. Nutrient dynamics in relation to surface–subsurface hydrological exchange in a groundwater fed chalk stream. J Hydrol. 2016;330:84–100.
Rovelli L, Attard KM, Binley A, Heppell CM, Stahl H, Trimmer M et al. Reach‐scale river metabolism across contrasting sub‐catchment geologies: effect of light and hydrology. Limnol Oceanogr. doi.org/10.1002/lno.10619. 2017.
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6 https://doi.org/10.1186/2049-2618-2-6
Pires AC, Cleary DF, Almeida A, Cunha A, Dealtry S, Mendonça-Hagler LC, et al. Denaturing gradient gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. Appl Environ Microbiol. 2012;78:5520–8.
Shen LD, Wu HS, Gao ZQ, Liu X, Li J. Comparison of community structures of Candidatus Methylomirabilis oxyfera-like bacteria of NC10 phylum in different freshwater habitats. Sci Rep. 2016;6:25647. 25610.21038/srep25647
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Muyzer G, de Waal E, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700.
Cadillo-Quiroz H, Bräuer S, Yashiro E, Sun C, Yavitt J, Zinder S. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ Microbiol. 2006;8:1428–40.
Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten M, Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol. 2009;75:3656–62.
Luesken FA, Zhu B, van Alen TA, Butler MK, Diaz MR, Song B, et al. pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol. 2011;77:3877–80.
Arshad A, Speth DR, de Graaf RM, Op den Camp HJ, Jetten MS, Welte CU. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by methanoperedens-like Archaea. Front Microbiol. 2015;6:1423 https://doi.org/10.3389/fmicb.2015.01423
Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531.
Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;63:183–229.
Op den Camp HJ, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, et al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep. 2009;1:293–306.
Huang Y, Zeng Y, Lu H, Feng H, Zeng Y, Koblížek M. Novel acsF gene primers revealed a diverse phototrophic bacterial population, including gemmatimonadetes, in Lake Taihu (China). Appl Environ Microbiol. 2016;82:5587–94.
Kojima H, Tsutsumi M, Ishikawa K, Iwata T, Mussmann M, Fukui M. Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol. 2012;35:233–8.
Ding ZW, Ding J, Fu L, Zhang F, Zeng RJ. Simultaneous enrichment of denitrifying methanotrophs and anammox bacteria. Appl Microbiol Biotechnol. 2014;98:10211–21.
Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ Microbiol Rep. 2009;1:377–84.
Zhu B, van Dijk G, Fritz C, Smolders AJ, Pol A, Jetten MS, et al. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl Environ Microbiol. 2012;78:8657–65.
Rasigraf O, Kool DM, Jetten MS, Sinninghe Damsté JS, Ettwig KF. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 2014;80:2451–60.
Shelley F, Grey J, Trimmer M. Widespread methanotrophic primary production in lowland chalk rivers. Proc Biol Sci. 2014;281:20132854.
Ryan P. Environmental and low temperature geochemistry. New York, NY: John Wiley and Sons; 2014.
Shelley F, Ings N, Hildrew A, Trimmer M, Grey J. Bringing methanotrophy in rivers out of the shadows. Limnol Oceanogr. 2017b;62:2345–59.
He Z, Zhang Q, Feng Y, Luo H, Pan X, Gadd GM. Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci Total Environ. 2018;610-611:759–68.
Lüke C, Frenzel P. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl Environ Microbiol. 2011;77:6305–9.
Kits KD, Klotz MG, Stein LY. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 2015;17:3219–32.
Padilla CC, Bertagnolli AD, Bristow LA, Sarode N, Glass JB, Thamdrup B et al. Metagenomic binning recovers a transcriptionally active gammaproteobacterium linking methanotrophy to partial denitrification in an anoxic oxygen minimum zone. Front Mar Sci. https://doi.org/10.3389/fmars.2017.00023. 2017.
Acknowledgements
This study was supported by the Natural Science Foundation of China (No. 41501261), the China Scholarship Council (CSC), the Natural Science Foundation of Jiangsu Province (No. SBK2015040035), and Queen Mary University of London. We acknowledge the land owners for granting us river access and also thank Ian Sanders and Felicity Shelley for technical and field work assistance and Chloe Economou and Monika Struebig for help with RNA work. We are also grateful to Alan Hildrew for his valuable suggestions on improving our manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shen, Ld., Ouyang, L., Zhu, Y. et al. Active pathways of anaerobic methane oxidation across contrasting riverbeds. ISME J 13, 752–766 (2019). https://doi.org/10.1038/s41396-018-0302-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0302-y
This article is cited by
-
High methane ebullition throughout one year in a regulated central European stream
Scientific Reports (2024)
-
Vertical distribution of Candidatus Methylomirabilis and Methanoperedens in agricultural soils
Applied Microbiology and Biotechnology (2024)
-
Nitrate-Dependent Anaerobic Methane Oxidation in Subtropical Mangrove Soils and Environmental Implication
Wetlands (2023)
-
Detection and Quantification of Candidatus Methanoperedens-Like Archaea in Freshwater Wetland Soils
Microbial Ecology (2023)
-
Water level of inland saline wetlands with implications for CO2 and CH4 fluxes during the autumn freeze–thaw period in Northeast China
Environmental Science and Pollution Research (2023)