Abstract
Information on how the oral microbiome develops during early childhood and how external factors influence this ecological process is scarce. We used high-throughput sequencing to characterize bacterial composition in saliva samples collected at 3, 6, 12, 24 months and 7 years of age in 90 longitudinally followed children, for whom clinical, dietary and health data were collected. Bacterial composition patterns changed through time, starting with “early colonizers”, including Streptococcus and Veillonella; other bacterial genera such as Neisseria settled after 1 or 2 years of age. Dental caries development was associated with diverging microbial composition through time. Streptococcus cristatus appeared to be associated with increased risk of developing tooth decay and its role as potential biomarker of the disease should be studied with species-specific probes. Infants born by C-section had initially skewed bacterial content compared with vaginally delivered infants, but this was recovered with age. Shorter breastfeeding habits and antibiotic treatment during the first 2 years of age were associated with a distinct bacterial composition at later age. The findings presented describe oral microbiota development as an ecological succession where altered colonization pattern during the first year of life may have long-term consequences for child´s oral and systemic health.
Similar content being viewed by others
Introduction
The development and structure of the neonatal microbiome have been partially elucidated, with a main focus on the microbial population inhabiting the lower intestinal tract, while information about the oral cavity colonization following delivery is still limited [1]. As yet, no published longitudinal studies have characterized oral microbiota development during infancy and childhood with culture independent next-generation sequencing methodologies, particularly in association with tooth decay.
It is believed that by production and excretion of metabolic products of pioneer colonizers (including facultative anaerobes Streptococcus and Actinomyces), acquired at birth and the following hours, the environment can be altered, thus benefiting and selecting the growth of other species (including more strictly anaerobic genera like Veillonella and Fusobacteria) [1, 2]. As the baby grows, microbial communities evolve and increase in microbial diversity [3, 4]. During this period, the oral microbiota is characterized by high variability and current knowledge indicates that it reaches adult-like stability around 2 years of age [1].
Most evidence available today shows that the early oral environment is strongly shaped by the mother [2, 5, 6] and maternal oral microbiota has been proposed to colonize the placenta [7] where it could influence foetal immune tolerance towards the mother's microbiome [8]. Further transition into a more mature and complex microbial ecosystem is mainly influenced by the external environment, as well as vertical transmission from the parents [2, 5, 9, 10]. An essential question is to identify which factors and at what time point they can influence the progression of microbial colonization. Previous studies of the lower gastrointestinal tract microbiota have reported that the gut microbiota of infants delivered by caesarean section (C-section) was mainly colonized by skin bacteria, had lower numbers of Bifidobacterium and Bacteroides species and were more often colonized with Clostridium difficile in comparison with vaginally born infants [11, 12]. However, research regarding the influence of delivery mode on the early oral microbiota development, by using next-generation sequencing on longitudinal samples, has not yet been reported.
Breast milk has long been considered a superior food for infants, increasing resistance to infections, providing nutrition and being a source of bacteria (106 bacterial cells/ml), who serve as inoculum for the newborn [13,14,15,16]. The genus Streptococcus is one of the dominant bacterial groups found in human milk [13, 15] and various species, including Streptococcus salivarius, are frequently found in the infant oral cavity [17]. The metabolic products derived from Streptococcus species from the dietary oligosaccharides in breast milk might pave the way for the establishment of other microorganisms in the oral cavity, thus influencing attachment and growth of selected bacteria [1, 2, 18,19,20,21]. However, the longitudinal impact of these initial colonizers on the oral ecosystem and the influence of breastfeeding habits on children’s oral and systemic health are widely unknown and deserve to be investigated.
Knowledge about the effect of other external factors like antibiotic use, especially at an early age, on subsequent microbiome development is also scarce. In children, long-term alterations of the gut microbiome as a consequence of early antibiotic administration have been described and proposed to have negative effects for systemic health, including obesity and allergy [22, 23]. However, the long-term effect of antibiotic use for children’s oral microbiota is currently unknown.
An important consequence of oral microbiome development for health is the protection against tooth decay (dental caries), considered among the most prevalent diseases worldwide [24]. Tooth decay is caused by an interaction between acidogenic bacteria, a carbohydrate substrate and host susceptibility, leading to bacterial dysbiosis and demineralization of tooth tissue [4, 25]. The acid-tolerant bacterial species Streptococcus mutans is recognized to be an important pathogen in dental caries, [26, 27] and its early presence in edentulous children (from 3 months of age), is suggesting that the soft tissue may play a role as a reservoir for oral pathogenic microorganisms [3, 28]. Given that early colonization with cariogenic microorganisms has been associated with higher caries incidence [26], microbiological studies in longitudinal samples through early childhood may reveal those bacteria increasing caries risk that could be used as early diagnostic biomarkers. This could also provide important information for active and passive immunization strategies against oral diseases [29]. Moreover, an unhealthy oral microbiome can have important effects beyond the oral cavity, including elevated cardiovascular risk [30, 31]. For instance, in vitro studies have demonstrated the ability of periodontal bacteria to increase the probability of thrombus formation, which could lead to ischaemic cardiovascular events [32, 33]. Therefore, it is of interest to understand the colonization patterns of oral commensals during childhood and the potential benign effect of oral bacteria in preventing oral and systemic diseases, including microorganisms, which have been associated with health conditions [34, 35].
A more detailed understanding of oral microbial communities development in health and disease fundamental and the use of high-throughput sequencing techniques now allow exploring microbial composition and diversity in low volume oral samples to an unprecedented level of detail [36], in comparison with culturing or early molecular methodologies. In this study, we aimed to address the temporal evolution and maturation of the oral microbial ecosystem during infancy and childhood and its relation to delivery mode, breastfeeding habits, antibiotic use and dental caries status, in longitudinally collected oral samples in 90 children followed from birth to 7 years of age.
Methods
Sample collection and study design
The infants included in the study were part of a larger randomized double-blind trial in Sweden between 2001 and 2003 evaluating the potential allergy prevention effect of probiotic Lactobacillus reuteri ATCC 55730 until 2 and 7 years of age [37, 38]. Among the 188 infants completing the original study, longitudinal salivary samples were collected in 90 children. The participants were instructed not to eat or drink for 2 h preceding the sampling. Non-stimulated saliva samples at 3, 6, 12 and 24 months of age were collected from the buccal cavity, using a hand pump (Nalgene #6131, ThermoFisher, Stockholm, Sweden) connected to a thin plastic tube and immediately frozen and kept at −80 °C. At 7 years of age, paraffin-stimulated whole saliva was collected (≈3 ml) in a sterile test tube and immediately frozen at −80 °C. By 9 years of age, the children were examined at public dental clinics at which the children received their regular dental care [39], and the caries status was evaluated. The oral examination included radiographs and the registration of manifest and initial caries lesions in the primary dentition according to Koch et al. and Alm et al. [40, 41].
Possible confounders, such as mode of delivery, breastfeeding habits (exclusive or partial breastfeeding), infant health and antibiotics use during the first 2 years of age were obtained from medical records and semi-structured questionnaires (see Table 1) [39]. In all, 91 and 80% of all children included were exclusively breastfed up to 1 and 3 months of age, respectively, whereas 97% were partially breastfed at 3 months of age. No infant received antibiotics before 1 month of age while 2% took antibiotics during the first 3 months of life.
The studies were approved by the Regional Ethics Committee for Human Research in Linköping, Sweden (Dnr 99323, M122–31 and M171–07, respectively). An informed consent was obtained from both parents before inclusion in the study. Written informed consent was also given by the parents or guardians before the dental examination.
DNA extraction
In all, 250 μl of each saliva sample were centrifuged at 15,000 g for 30 min and the pellet, together with 50 μl of the supernatant, was used for further analysis. DNA was isolated by MagNA Pure LC 2.0 Instrument (1996–2016 Roche Diagnostics, Barcelona, Spain), using MagNA Pure LC DNA Isolation Kit III for Bacteria and Fungi (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer’s instructions with an additional enzymatic lysis step with lysozyme (20 mg/ml, 37 °C, 60 min; Thermomixer comfort, Eppendorf, Hamburg, Germany), lysostaphin (2000 units/mg protein, 37 °C, 60 min; Sigma-Aldrich, Madrid, Spain) and mutanolysin (4000 units/mg protein, 37 °C, 60 min; Sigma-Aldrich). DNA was resuspended in 100 μl of elution buffer and frozen at −20 °C until further analysis.
16S rRNA gene amplification and sequencing
Prior to sequencing of 16S ribosomal RNA (rRNA) gene, extracted DNA was pre-amplified by using universal bacterial degenerate primers 8F—5ʹ-AGAGTTTGATCMTGGCTCAG-3ʹ and 926R—5ʹ-CCGTCAATTCMTTTRAGT-3ʹ, which encompass the hypervariable regions V1–V5 of the gene. This was performed using the high-fidelity AB-Gene DNA polymerase (Thermo Scientific, Waltham, Mass., USA) with an annealing temperature of 52 °C and 10 cycles, in order to minimize amplification biases [42]. The purification of PCR products was completed using Nucleofast 96 PCR filter plates (Macherey-Nagel, Düren, Germany).
An Illumina amplicon library was performed following the 16S rRNA gene Metagenomic Sequencing Library Preparation Illumina protocol (Part #15044223 Rev. A). The gene‐specific primer sequences used in this protocol were selected from Klindworth et al. [43], and target the 16S rRNA gene V3 and V4 regions, resulting in a single amplicon of approximately 460 bp. Overhang adapter sequences were used together with the primer pair sequences for compatibility with Illumina index and sequencing adapters. After 16S rRNA gene amplification, the DNA was sequenced on a MiSeq Sequencer according to manufacturer’s instructions (Illumina) using the 2 × 300 bp paired-end protocol. Sequences supporting the conclusions of this article are publicly available at the European Nucleotide Archive (ENA) database with the accession number PRJEB66628.
Bacterial load and Streptococcus dentisani measurements with quantitative PCR
Total bacterial load (number of bacterial cells per ml of saliva) and the presence of Streptococcus dentisani in saliva samples were measured by quantitative PCR. Amplifications were performed in duplicates on a LightCycler 480 Real-Time PCR System (Roche Technologies) by using annealing temperatures of 60 °C and 65 °C for total bacterial load and S. dentisani, respectively. Each reaction mixture of 10 mL was composed of SYBR Green PCR Master Mix (Roche), 0.5 mL of the specific primer (concentration 10 mmol/L), and 2 mL of DNA template. For S. dentisani, the forward primer was 5ʹ-GTAACCAACCGCCCAGAAGG-3ʹ and the reverse primer 5ʹ-CCGCTTTCGGACTCGATCA-3ʹ (Integrated DNA Technologies (IDT); San Diego, California, USA) targeting the carbamate kinase gene, and for total bacterial density measurements the universal forward and reverse primers were 5ʹ-GTGCCAGCMGCCGCGGTAA-3ʹ and 5ʹ-GCGTGGACTACCAGGGTATCT-3ʹ (IDT), respectively, targeting the bacterial 16S rRNA gene. The obtained Ct values were transformed in bacterial cell numbers by a standard curve calibrated by flow cytometry [13].
Bioinformatics and statistics
Only overlapping paired-end reads were used for analysis. A sequence quality assessment was carried out using the PRINSEQ program [44]. Sequences of <250 nucleotides in length were not considered; sequence end-trimming was performed by cutting out nucleotides with a mean quality of <30 in 20-bp windows. Chimeric 16S sequences were filtered out using USEARCH program [45].
Obtained sequences were taxonomically classified by the RDP classifier [46] where reads were assigned a phylum, class, family and genus and phylogenetic ranks were allocated when scores exceeded an 0.8 confidence threshold. Operational taxonomic units (OTUs) were generated by using CD-HIT OTU picking with 97% of similarity [47]. Human oral microbiome database (HOMID) was used as a reference database for OTU assignment [48]. For the Streptococci species analyses, sequences were clustered into OTUs at 100% similarity by BLAST analysis [49] and >350 bp alignment length, against the RDP database [50]. A few species appeared to be identical in the sequenced region, namely Streptococcus infantis, S. mitis and S. dentisani, and could not be distinguished from each other.
α-Diversity analyses (presented here as Shannon and Chao1 indices), were utilized to estimate samples’ diversity and richness at the 97% OTU level using the R-package Vegan [51]. Constrained correspondence analysis (CCA, a.k.a. canonical correspondence analysis) is a statistic tool used to emphasize variation, taking advantage of the fact that the factor provided can explain part of the total variability, and bring out strong patterns in a dataset. This analysis was performed by R software ade4 package [52] using the function CCA, which is based on Chi-squared distances. Adonis tests were done with the R library ‘vegan’ [51]. It performs a permutational multivariate analysis of variance using distance matrices and fitting linear models to them. The test allows modelling the whole compositional variability at once by taking into account different sources of variation, as well as interactions between them as it is defined in a linear model.
Linear discriminant analysis effect size (LEfSe), a method for biomarker discovery on the online interface Galaxy (http://huttenhower.sph.harvard.edu) [53], was used to detect the taxa, at both genus and OTU level, characterizing the populations of caries-free and caries-active children.
Statistical analyses were performed in R version 3.2.2 and GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA, Version 6.1f), where p < 0.05 was considered significant. Specific statistical tests (including Mann–Whitney U-test for nonparametric comparisons) are stated in figure legends. When comparing the frequencies of different bacterial taxa between groups (e.g., caries-free and caries-experienced children), the balanced proportions of confounding factors, including breastfeeding length, mode of delivery and antibiotic intake, were checked by chi-square test, and nonsignificant differences between the groups were found.
Findings and discussion
After quality filtering, 34,794,056 sequences were obtained, with an average of 93,532 ± 3480 (SEM) sequences per sample.
Bacterial load, richness and diversity through time
Bacterial diversity and richness increased through time, reaching nearly 550 OTUs at 7 years of age with a Shannon diversity index of approximately 2.4 (Fig. 1). The delivery mode and partial breastfeeding habits until 12 months of age did not have an impact on species richness (Fig. 1a, b). However, bacterial diversity appeared to be higher in C-section delivered infants at 12 months of age (Fig. 1a) and at 2 years of age in children not being breastfed through 12 months of age (Fig. 1b).
Species richness and diversity of total microbiota in infant saliva samples. Bacterial richness and diversity (here presented by Chao1 and Shannon indices), obtained at different time points from birth to 7 years of age, were determined by 16S rRNA Illumina sequencing and OTU clustering at 97% sequence identity. a shows species richness and diversity in infants delivered vaginally (VD) or by caesarean section (C-section) at 3 months (NVD = 62; NCS = 11), 6 months (NVD = 72; NCS = 11), 12 months (NVD = 59; NCS = 10), 24 months (NVD = 56; NCS = 10) and 7 years of age (NVD = 68; NCS = 12). b represents species diversity and richness in infants breastfed for 12 months (BF) and in infants breastfed for less than 6 months of age (nBF). Analyzed samples were collected at 12 months (NnBF = 52; NBF = 17), 24 months (NnBF = 50; NBF = 16), and 7 years of age (NnBF = 59; NBF = 21). c shows species richness and diversity in children developing caries (CA) and children staying caries-free (CF) during the first 9 years of life. Saliva samples were collected at 3 months (NCF = 40; NCA = 26), 6 months (NCF = 43; NCA = 31), 12 months (NCF = 37; NCA = 26), 24 months (NCF = 35; NCA = 24) and 7 years of age (NCF = 45; NCA = 30). Data are presented with means and standard errors. (*p < 0.05; Mann–Whitney U-test)
Oral development, including the emergence of teeth, was accompanied by a steady increase in diversity and richness of the oral microbiome in this study, especially between 1 and 2 years of age. Interestingly, bacterial diversity at 2 years of age (Fig. 1b), appears to be higher in children, which abandoned breastfeeding before 12 months of age. Although this has not been studied before in oral microbiota, a similar trend was observed in gut microbiota analyses where children not being breastfed had higher microbial diversity [54,55,56], probably due to earlier introduction of solid food. Our results agree with a scenario in which following delivery, the oral cavity gets exposed to the environment, triggering the initiation of microbial colonization through diet, vertical transmission from parents and horizontal transmission from caregivers and siblings, thus increasing the bacterial diversity [57, 58].
In order to determine the development of bacterial density through infancy, we measured total bacterial load (Fig. SI1) in saliva samples. Although there were no differences regarding delivery mode (Fig. SI1a) and breastfeeding habits (Fig. SI1b), the density of bacteria increased significantly with age, probably reflecting the influence of environmental interactions and the emergence of teeth. Interestingly, bacterial density at each time point appeared to fall within two groups (low or high), and this bimodal distribution was maintained through time for each individual. This pattern could not be attributed to caries status, allergy development, mode of delivery, feeding habits, antibiotics intake or probiotic administration (data not shown). In the future, it would be interesting to determine whether the physicochemical properties of saliva may influence cell density.
Bacterial composition during infancy
When bacterial composition was analyzed for all samples through child development, clear changes emerged through time (Fig. SI2). Streptococci dominated salivary samples at all times. They were particularly high in proportion during the first months of age, and their decrease was accompanied by a rise in other genera. These general patterns were influenced by several perinatal and postnatal factors.
The influence of delivery mode and breastfeeding durations
Bacterial species composition development was influenced by delivery mode and breastfeeding habits (Fig. 2a, b), but not by L. reuteri supplementation during the first year of age (data not shown). The impact of delivery mode was reflected in differences in bacterial composition at 3 and 6 months of age (Fig. 2a, p = 0.001, CCA analysis), followed by convergent microbial patterns at later age. Only the genus Haemophilus was found to be significantly more abundant (p = 0.047) at 7 years of age in children delivered by C-section (Fig. SI3). Thus, with the exception of this genus, no further colonizers were found to be significantly different between vaginally delivered and C-section delivered infants (Fig. SI3). This could be due to infant delivery mode affecting the direct transmission of initial bacteria from mother to newborn, having a short-term effect. This finding is in line with previous studies [59] where the Human Oral Microbe Identification Microarray was used, showing that microbial oral colonization in 3-month-old infants delivered vaginally and those delivered by C-section was different. Similar findings of an early impact, but also more long-term effects [11, 60], have been reported for the microbiota of the lower gastrointestinal tract [12, 61]. When a multivariate analysis was performed including time, breastfeeding length and caries status as confounding factors, the effect of delivery mode on microbiota composition was no longer significant. Given that a significant breastfeeding length-delivery mode interaction was detected (p = 0.026), part of the observed differences between children born by vaginal delivery and C-section can be due to the effect of breastfeeding.
Salivary microbiota patterns through children’s development. Constrained correspondence analysis (CCA), here used to emphasize variations in microbiota species-level profiles, show the relationship between groups in total microbiota composition at different time points. The percentage of variation explained by constrained correspondence components is indicated on the axes. a Microbial pattern differences in saliva from infants delivered vaginally (VD) or by caesarean section (CS), p = 0.0016, at 3 months (NVD = 62; NCS = 11), 6 months (NVD = 72; NCS = 11), 12 months (NVD = 59; NCS = 10), 24 months (NVD = 56; NCS = 10) and 7 years of age (NVD = 68; NCS = 12). b Showing microbial composition pattern differences in infants who were breastfed for 12 months (BF) and in infants breastfed (nBF) for less than 6 months (p = 0.0017). The numbers of children were: 12 months (NnBF = 52; NBF = 17), 24 months (NnBF = 50; NBF = 16) and 7 years of age (NnBF = 59; NBF = 21). c Microbial composition patterns in children developing caries (CA) and children staying caries-free (CF) during the first 9 years of life (p = 0.0018). Saliva samples were collected at 3 months (NCF = 40; NCA = 26), 6 months (NCF = 43; NCA = 31), 12 months (NCF = 37; NCA = 26), 24 months (NCF = 35; NCA = 24) and 7 years of age (NCF = 45; NCA = 30). Numbers accompanying the variables (delivery mode, breastfeeding and caries onset) are representing the time points plotted. p-Values for CCA plots were determined by Adonis analysis (a nonparametric statistical method, R package Vegan) and significant values indicate that the factor provided can explain part of the total variability
The influence of partial compared with no breastfeeding until 12 months of age did appear to have a long-term effect, as evidenced by a divergent oral bacterial composition at 24 months and 7 years of age (Fig. 2b, p = 0.002) while bacterial colonization at early age appeared to be similar. This could be due to the fact that the majority of the infants in this cohort were breastfed during their first months of life (see Table 1). A multivariate analysis revealed that the significant effect of breastfeeding on microbiota composition was maintained even after removing the effect of caries status, time and mode of delivery as confounding factors (p = 0.036). Further work should therefore address the impact of formula feeding on microbiome development as findings presented here suggest that variations in the initial oral microbial communities may result in differences in the bacterial succession patterns that persist over time, analogous to the impact of early disturbance in ecological successions [62].
Microbial colonization patterns
Dominant bacterial genera (present at >1%), which inhabited the oral cavity during the first 3–6 months, here called “Early colonizers”, included Streptococcus, Veillonella and Lactobacillus spp. (Fig. 3a). The most frequent bacterium of the oral cavity in the current study was Streptococcus, and children being breastfed until 12 months of age appeared to have higher abundance of this genus at one year of age (p = 0.005). This finding is consistent with other reports [3, 63] and Streptococcus has been found to be one of the dominant bacterial groups in human breast milk [13, 16]. Aging of the children was associated with lower levels of Streptococcus, although the decrease tended to be more notable in children abandoning breastfeeding before 12 months of age. This indicated that settlement of this genus is favoured by breast milk, either by direct transmission or by an appropriate nutrient supply [13, 64]. Moreover, this pioneer is often found in the oral cavity of the neonate because of its ability to adhere to and colonize the mucosal surface lining [2]. The metabolic products (such as lactic acid) derived from Streptococcus species from the dietary oligosaccharides in breast milk might pave the way for the establishment of other microorganisms in the oral cavity, including bacterial genera like Veillonella [1, 21]. Veillonella, here ranging between 2 and 8% of total abundance with significantly higher levels at 7 years in children keeping breastfeeding until 12 months of age (p = 0.037), is another bacterial genus commonly encountered in breast milk [65, 66]. This genus requires organic acids as carbon source and therefore its presence is likely favoured by the high levels of lactate derived from lactose fermentation, which this genus will transform to propionate and acetate [67]. An important lactose fermenter is obviously Lactobacillus, which in the oral cavity might be acquired by the neonate during vaginal delivery, as this genus is highly abundant in vaginal microbiota, [68] but also through breastfeeding since breast milk has been proposed to favour the growth of vaginally acquired bacteria [61, 67, 69]. In the current study, no differences in Lactobacillus abundance were found between children being breastfed up to one year of age or not (Fig. 3a) and neither between vaginally delivered and C-section infants (Fig. SI3). Among the components of human breast milk, oligosaccharides are thought to directly influence the gut microbial composition and to enrich bacterial functions associated to carbohydrate consumption and biosynthesis of amino acids and vitamins [55, 70] and a similar process may be taking place in the oral cavity. Early commensals of the oral cavity are likely having an ecological advantage over those arriving later and may promote the change of the environment through the production and excretion of products of their metabolism, thus benefitting the growth of further oral bacterial communities. This process of microbial succession and increasing diversity, promoted by breastfeeding, could lead to subsequent formation of complex and steadier microbial communities, as proposed for gut microbiota [71].
Microbiota composition of dominant bacterial genera in children being or not being breastfed until 12 months of age. a Genera classified as early colonizers. b Genera considered to “constantly increase” are already present at 3–6 months of age at >1% frequency, and are increasing in relative proportion with time. c Bacterial genera considered as “Late colonizers”, are defined as those present at 3–6 months of age below 1% relative abundance, which undergo a significant increase after 12 months of age. Plots are showing the relative abundance of dominant bacterial genera, as determined by Illumina sequencing of 16S rRNA gene, in saliva samples collected at 3 months, 6 months, 12 months (NnBF = 52; NBF = 17), 24 months (NnBF = 50; NBF = 16) and 7 years (NnBF = 59; NBF = 21) of age. Asterisks indicate cases where p < 0.05 by both Mann–Whitney U-test and Wilcoxon analysis
Bacterial genera Gemella, Granulicatella, Haemophilus and Rothia, here defined as “constant colonizers” (Fig. 3b), were present already at 3 and 6 months of age with >1% of abundance, and their abundance increased with time. Gemella and Granulicatella are considered as common dental plaque inhabitants [72] and were found to increase in abundance through age, ranging from 5 to 10 and 2 to 8%, respectively. It is likely that the initiation of teeth eruption, starting around 6–8 months postnatally, creates new ecological niches in the oral cavity, giving rise to new adhesion surfaces, thus favouring their further colonization.
A third set of microorganisms were “late colonizers” and included Actinomyces, Porphyromonas, Abiotrophia and Neisseria, which became dominant in the oral cavity at a later stage, approximately after the first year of life (Fig. 3c). Thus, the data suggest that the acquisition or dominance of each bacteria may occur optimally only at certain ages. Breastfeeding until 12 months of age was associated with significantly lower levels of Actinomyces (p = 0.044) at 7 years of age and Porphyromonas (p = 0.049) and Neisseria (p = 0.028) at 12 months and 24 months of age, respectively. Porphyromonas, more specifically Porphyromonas gingivalis, is a Gram-negative oral anaerobe involved in the pathogenesis of periodontitis, an inflammatory disease that destroys the tooth tissue and may lead to tooth loss [73]. The results are indicating that children being breastfed by 12 months of age, as compared with children no longer breastfed, have significantly lower abundance of this genus at 1 year of age. However, species-level taxonomic analysis revealed that 100% of Porphyromonads sequences correspond to Porphyromonas catoniae during the first 12 months of age. At 2 years, P. gingivalis appeared at 9% of the total, whereas P. catoniae accounted for 91% of the sequence reads. At 7 years of age, the proportions were 86.5% for P. catoniae and 13.4% P. gingivalis. Thus, an association between reduced breastfeeding length and risk of gum disease is uncertain. Neisseria, a common bacterial community member of the healthy human mouth [74], was found to be more abundant in children not being breastfed until 12 months of age, in line with previous research where species belonging to this genus were found more frequently in children being formula-fed [75]. Thus, breast milk had a long-term effect on oral microbiota composition, but this altered microbiota could not always be linked to healthy or disease-associated communities, and further work should study the long-term consequences for the child’s oral and systemic health. Beside the potential health effect, the results presented here are suggesting that the transmission of bacteria from breast milk and the nutrients supplied by it at this critical time point in infantʼs development, could affect the colonization window of specific bacterial genera, and depending on delivery mode and breastfeeding duration, this may lead to disturbances in the oral microbial succession patterns that persist over time.
The effect of antibiotics intake on microbiota development
The clinical data of this cohort allowed us to assess the influence of antibiotics intake in early life (first and second year) on developing microbiota. The antibiotics courses given were mainly due to early otitis media (in 89% of cases) and included Amoxicillin (34 % of cases) and Phenoxymethylpenicillin (42 % of cases) (Table S1). Upon comparing the microbial succession in children who did or did not take antibiotics during the first 2 years of life, significantly divergent colonizations were observed at 24 months and 7 years of age, whereas bacterial composition at earlier time points were overlapping in children treated with antibiotics (Fig. 4a). Multivariate analyses were also performed, considering the effect of time and different confounding factors on microbiota composition. Antibiotic use had a significant effect on microbiota composition once the effects of caries status and time were removed (p = 0.05) and a significant antibiotic by time interaction was found (p = 0.008). There was a lower effect of antibiotics on microbiota composition (p = 0.067) once breastfeeding length was included in the analysis, suggesting that part of the significance is due to the strong effect of breastfeeding on microbiota composition.
Accumulative antibiotics effect on salivary microbiota development. a shows salivary microbiota patterns in children treated with antibiotics during the first 2 years of life and children not taking any antibiotics during the first 2 years of life; p = 0.017. p values for CCA plots were determined by Adonis analysis (a nonparametric statistical method, R package Vegan) and indicate that the factor provided can explain part of the total variability. b represents the influence of antibiotics intake on bacterial species distribution at 24 months and 7 years of age, here represented with a Venn’s diagram. The numbers indicate unique species found in children not treated with antibiotics (purple) and children treated with antibiotics (grey) and OTUs differentially distributed are stated in the tables. OTUs presented were filtered according to sequence length (>300 bp) and identity (>97% nucleotide similarity). Analyzed sample sizes were: 3 months (NNO = 28; NYES = 11), 6 months (NNO = 33; NYES = 13), 12 months (NNO = 29; NYES = 11), 24 months (NNO = 26; NYES = 10) and 7 years (NNO = 35; NYES = 11); NO = no antibiotics intake for the first 2 years of age; YES = antibiotic consumption during the first 2 years of age
By comparing the most dominant genera (>1% of total microbiota) present in these two groups, the genus Granulicatella was higher in abundance at 24 months of age (p = 0.003) in children not taking antibiotics while Prevotella (p = 0.020) was more prevalent at 7 years of age in children treated with antibiotics in early life. The data suggest that the abundance of commensal genera such as Granulicatella [72] may be disturbed by antibiotics use, whereas the presence of other genera, like Prevotella, which has been associated with several oral diseases [76], may be favoured.
In order to obtain deeper insight of microbiota alterations upon antibiotics intake, the microbial composition was assessed at species-level OTUs (Fig. 4b). The analysis revealed a high number of bacteria uniquely present in children that were treated with antibiotics more than once during the first two years of life including several Actinomyces species at 2 and 7 years of age. Moreover, the presence of species belonging to Fusobacterium, Veillonella and Lactobacillus was also associated with antibiotics intake during the first 2 years of life in our cohort. The fact that Veillonella spp use organic acids as their only carbon source strongly suggests that the oral microbiota of those children is more acidogenic. On the contrary, Neisseria and Streptococcus mitis/dentisani, were present in our samples at significantly higher levels in 7-year-old children that did not take antibiotics. Thus, although a divergent microbiota does not necessarily imply a negative effect for health, most significant changes in microbial composition detected in the current study as a consequence of antibiotic administration, have previously been associated with oral diseases ([35, 36, 77, 78–81]) and future studies will need to specifically address whether antibiotic use during infanthood has an effect on oral health.
It is of course possible that the divergent microbial succession patterns observed at 7 years of age might be affected by further antibiotics courses and other influencing factors, occurring during the remaining 5 years. However, given that the first years of age appear to represent a crucial period of microbiota development and immune system modulation and that early changes in ecological successions are those with the largest impact on community development [62], it is important to consider that early antibiotic treatment can have long-term consequences for microbiota development. It has to be emphasized that in adults, the original salivary microbial composition appears to be restored after antibiotic use [82], suggesting resilience of the oral microbiome; in children, long-term alterations of the gut microbiome, as a consequence of early antibiotic administration, have been proposed to have negative effects for systemic health, including obesity and allergy [22, 23]. Thus, the impact of early intake of antibiotics for human health deserves consideration.
Oral microbiota in health and disease
Caries development did not appear to be related with bacterial diversity (Fig. 1c) or bacterial load (Fig. SI1c) during the first 7 years of life. Although there were no differences between children staying caries-free and children developing caries at age 9, the density of bacteria was increasing significantly with age, probably reflecting the influence of environmental interactions and the emergence of teeth. The overall species richness was higher in children that remained caries-free by 9 years of age, but the difference was not significant (Fig. 1c). However, the potential association of lower bacterial diversity to caries risk should be further studied, as a lower bacterial diversity has been associated to caries in cross sectional studies [83]. A factor reducing the possible association of caries status to diversity could be the use of saliva samples, which provide a good representation of overall oral microbial diversity but may not fully correlate with bacterial composition at the tooth biofilm, where the disease takes place [84].
Caries development at 9 years of age was preceded by divergent bacterial composition at 24 months of age, reaching the maximum at 7 years (Fig. 2c). At early age, no differences between caries-experienced and caries-free children were detected, suggesting that the colonization patterns and ecological factors favouring caries development are associated with later age. A critical period may include the eruption of primary incisors, primary molars and permanent first molars, where cariogenic bacteria like Mutans streptococci can adhere through glucan binding proteins [85]. Although these caries-linked species are considered associated to hard tissues, there are studies suggesting that they can be acquired at any time from under 6 months (prior to first tooth eruption) to over 3 years of age [86, 87]. Taken together, the data here suggest that different bacterial colonization patterns were present between caries-free children and children that developed caries, however, they were significant only after the second year of age.
Bacterial composition and caries development
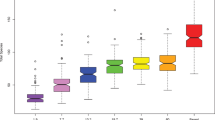
As no significant differences were observed between caries-free and caries-active children at the genus taxonomic level (Fig. SI4) and given that the genus Streptococcus was highly abundant in the infants’ oral cavity, it was of great interest to investigate if there were any specific Streptococci species associated with caries development in the cohort. The genus Streptococcus comprises a large number of species that can have positive effects on human health and some of them have started to be used as probiotics in oral diseases [84]. The OTUs found corresponded to S. mitis/infantis/dentisani (identical in the sequenced 16S rRNA region), S. salivarius, S. sanguinis, S. lactarius, S. cristatus and S. mutans (Fig. 5). S. mitis/infantis/dentisani were the most prevalent OTUs (ranging from 75 to 85%) and no difference was found between the children who did or did not develop caries at 9 years of age. S. infantis belongs to the Streptococcus mitis group [89] and has been associated with oral health as it significantly decreases during caries progression in the young permanent dentition [90]. S. dentisani is a bacterial species associated with good oral health and it has been isolated from caries-free individuals [35]. Because of the high sequence similarity within the Streptococcus genus PCR-amplified region used for Illumina sequencing, we could not distinguish which 16S rRNA reads belonged to S. mitis, S. infantis or S. dentisani. To clarify this, quantitative PCR (qPCR) amplification with S. dentisani-specific primers was performed in order to determine the acquisition of this species through age. The quantities of S. dentisani were undetectable by qPCR during the first year of age, suggesting that the colonization of this species might be dependent of teeth eruption. This is in agreement with its normal association with dental plaque [35]. The levels of S. dentisani were higher in children remaining caries-free at 9 years of age in comparison with caries-experienced children, but the difference was not significant (Fig. SI5).
Relative abundance of most prevalent Streptococci species found in saliva samples of children staying caries-free and children developing caries during the first 9 years of age. Plots represent average relative abundance of Streptococci through time. Taxonomy assignments were performed with RDP classifier at 100% nucleotide identity. All data are presented as means with standard errors. Asterisks indicate cases where p < 0.05 by both Mann–Whitney U-test and Wilcoxon analysis
Streptococcus salivarius was another commonly found species in children’s saliva (Fig. 5). Its abundance was highest at 3 months of age, ranging between 10 and 15% of the total streptococcal species, and decreasing steadily through time, likely opposing teeth eruption. This pioneer colonizer and a prominent member of the oral microbiota of the healthy mouth has been detected hours after birth because of its unique ability to adhere and colonize tongue and cheek mucosa [58]. Although S. salivarius has been intended for use as a probiotic targeting the oral cavity [91], no differences in abundance levels of this species through age were discovered between children who did or did not develop caries at 9 years of age, perhaps due to its absence from dental plaque [35]. Streptococcus lactarius was another species encountered in infant´s saliva, predominantly at 3 and 6 months of age, to later decrease and even disappear. This species was isolated from breast milk of healthy mothers [92], explaining its high abundance in early age when the majority of the children in this cohort were breastfed. Given the long-term impact of breastfeeding for microbiota development (Fig. 2b), it is plausible that early colonization with S. lactarius, acquired from motherʼs breast milk, could benefit later colonization by other beneficial microbial species. However, the potential role of S. lactarius in health and disease has not been evaluated to date.
Colonization of Streptococcus sanguinis started between 6 and 12 months of age and followed a similar pattern of development between children who did and did not develop caries. This species is believed to play a benign role in the oral cavity and it has been described to colonize in association to tooth emergence, at a median age of 9 months [93]. Moreover, S. sanguinis is recognized for its antagonistic role in dental caries as it may compete with cariogenic mutans streptococci for colonization sites on tooth surfaces [93]. Interestingly, although at very low levels, the cariogenic S. mutans was detected in the oral cavity of the infants already at an early age, possibly acquired through their mothers as shown before, [85] with a trend of significantly higher levels at 7 years (p = 0.06) in children developing caries. This is in line with previous studies where proportions of S. mutans in saliva were higher in children with caries when compared with those who stayed caries-free [4]. Thus, although this species is considered mainly an inhabitant of hard tissues, our data show that it can be detected before tooth eruption and therefore the oral health of mothers and caretakers during infancy may play an important role in the transmission of this pathogen. However, S. mutans has also been detected in caries-free populations and not in all cases of childhood caries, suggesting that other species may be cariogenic pathogens [76, 85]. In this study, children developing caries had significantly higher abundance of Streptococcus cristatus already at 3 months (p = 0.026) and 24 months of age (p = 0.033), compared with the children that stayed caries-free until 9 years of age. Given that S. cristatus, among other species, has been associated with severe early childhood caries [27], even in the absence of Streptococcus mutans, its role as an important cariogenic species and potential caries risk biomarker should be further studied. Nevertheless, it must be emphasized that streptococci are extremely similar in their 16S rRNA gene sequence, particularly at the V3–V4 region analyzed in the current work, and therefore the suggested association of S. cristatus with caries development should be confirmed by species-specific methodology with higher discriminatory power, like qPCR with specific primers and probes [94].
If the association between S. cristatus and dental caries is confirmed, it must be born in mind that this species has been found to interrupt the formation of P. gingivalis biofilms by repressing the production of several virulence factors in this major periodontal pathogen [95]. In our dataset, a scatterplot of the relative frequencies of Porphyromonas and S. cristatus shows an L-shape (correlation p-value for the hyperbolic regression was p = 0.057), suggesting potential antagonistic behaviour (Fig. SI6), a feature that has been demonstrated in subgingival plaque samples from adults [96]. Given that most Porphyromonas sequences in our samples corresponded to P. catoniae (P. gingivalis accounted only for 13.4% of total Porphyromonas reads by 7 years of age), the potential antagonism between S. cristatus and P. gingivalis may not be apparent until a later age.
In addition, LefSe analyses were performed in order to examine potential biomarkers for early caries diagnosis. No specific group of species/genera at early age could be associated with caries development at 9 years of age (data not shown), suggesting that other ecological determinants including host interactions with microbiota, play a crucial role and should be integrated in caries risk assessment models [97, 98]. Interestingly, even though the supplementation with L. reuteri during the first year of life has been associated with reduced caries prevalence at 9 years of age [39], no differences in caries development related to this Lactobacilli could be detected in the present study. Given that some of the infants included in the study developed allergies during their early childhood (see Table 1), the groups were balanced according to allergy status and no relationship was found between allergies and caries onset. Even though mode of delivery and breastfeeding until 12 months of age have been shown to impact oral microbiota development in this study, no correlation between delivery mode or breastfeeding duration with dental caries could be detected. However, this could be due to low statistical power of the groups compared. Although microbiota composition clearly differed at 7 years of age between caries-free and caries-experienced children (Fig. 2c), the absence of robust individual biomarkers of caries risk at an earlier age underlines that microbial-based early diagnostic tests should not be based on single species, and new potential bacterial risk indicators should be identified [99], including S. cristatus as proposed above. Given the enormous inter- and intra-individual variability in bacterial composition at caries lesions [100], and the multi-factorial nature of oral diseases where microbial, environmental and host-associated variables are involved, a holistic, ecological approach to caries risk assessment where information about the host, the habits (including the diet and oral hygiene) and the microbes are integrated will likely provide a better estimate of caries prediction [97, 98, 101].
Conclusions
Only limited information is available on oral microbiome development in infants, and most studies have focused on taxonomic analysis. Thus, functional, metagenomic analyses are pending to fully understand the microbial contribution to oral health and disease [84]. Previous studies addressing oral microbiota development in early life have been hampered by retrospective approaches, small sample sizes, lack of deep sequence coverage, limited period of follow-up and analyses at single time points. The current study demonstrates that the infant’s oral cavity gets colonized by microorganisms in a timely manner, increasing in complexity with time. In general, the data presented in the current manuscript is consistent with a model where microbiota development follows an ecological succession [102].
In this scenario, several early colonizing species pave the way for the settlement of other microorganisms, which further expand microbial diversity towards a mature community, which is more robust and resilient to change, partly because of the developed immune tolerance [8]. The presence of several species (particularly S. cristatus) at an early age was associated in this study to a higher frequency of dental caries at 9 years of age. Therefore, these findings open the possibility to use this species, together with others identified in other studies, as potential biomarkers of caries risk. The oral cavity is a complex and heterogenous ecosystem with many variables influencing microbial composition and function. Several external factors appear to strongly influence microbiota development, including mode of delivery, which had a short-term effect, and others like breastfeeding length or antibiotic treatment, which appeared to have a long-term impact. It is interesting to note that, on the contrary, the oral microbiome composition in adults appears to be extremely resilient to antibiotic treatment [82]. This highlights that developmental milestones that are critical for oral microbiota succession occur in particular during infancy, and that an appropriate microbial colonization pattern can be instrumental for future health. Thus, microbial exposure, feeding habits and medical interventions during those initial and fragile stages may have a lifelong impact on general microbiome composition, and their potential consequences for human health should be carefully studied.
References
Gomez A, Nelson KE. The oral microbiome of children: development, disease, and implications beyond oral health. Microb Ecol. 2017;73:492–503.
Sampaio-Maia B, Monteiro-Silva F. Acquisition and maturation of oral microbiome throughout childhood: an update. Dent Res J (Isfahan). 2014;11:291–301.
Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, Meline BS, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503.
Lif Holgerson P, Öhman C, Rönnlund A, Johansson I, Könönen E, Könönen E, et al. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10:e0128534.
Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531.
Teng F, Yang F, Huang S, Bo C, Xu ZZ, Amir A, et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015;18:296–306.
Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237–65.
Zaura E, Nicu EA, Krom BP, Keijser BJF. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:85.
Hesselmar B, Sjöberg F, Saalman R, Åberg N, Adlerberth I, Wold AE. Pacifier cleaning practices and risk of allergy development. Pediatrics. 2013;131:1829–37.
Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458.
Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut. 2014;63:559–66.
Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21.
Boix-Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. 2016;7:492.
Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, et al. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10.
Fitzstevens JL, Smith KC, Hagadorn JI, Caimano MJ, Matson AP, Brownell EA. Systematic review of the human milk microbiota. Nutr Clin Pract. 2016;32:354–64.
Rodriguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr Int Rev J. 2014;5:779–84.
Carlsson J, Grahnen H, Jonsson G, Wikner S. Early establishment of Streptococcus salivarius in the mouths of infants. J Dent Res. 1970;49:415–8.
Danielsson Niemi L, Hernell O, Johansson I. Human milk compounds inhibiting adhesion of mutans Streptococci to host ligand-coated hydroxyapatite in vitro. Caries Res. 2009;43:171–8.
Sheedy JR, Wettenhall REH, Scalon D, Gooley PR, Lewis DP, MCGregor N, et al. Increased D-lactic acid intestinal bacteria in patients with chronic fatigue syndrome. Vivo (Brooklyn). 2009;23:621–8.
Aimutis WR. Bioactive properties of milk proteins with particular focus on anticariogenesis. J Nutr. 2004;134:989S–95S.
Wernersson J, Danielsson Niemi L, Einarson S, Hernell O, Johansson I. Effects of human milk on adhesion of Streptococcus mutans to saliva-coated hydroxyapatite in vitro. Caries Res. 2006;40:412–7.
Reynolds LA, Finlay BB. Early life factors that affect allergy development. Nat Rev Immunol. 2017;17:518–28. https://doi.org/10.1038/nri.2017.39
Ajslev TA, Andersen CS, Gamborg M, Sørensen TIA, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. 2011;35:522–9.
Petersen PE. The World Oral Health Report 2003: continuous improvement of oral health in the 21st century—the approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol. 2003;31:3–24.
Selwitz RH, Ismail AI, Pitts NB. Dental caries. Lancet. 2007;369:51–9.
Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Moore A, Hughes CV, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–97.
Tanner ACR, Mathney JMJ, Kent RL, Chalmers NI, Hughes CV, Loo CY, et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49:1464–74.
Nelun Barfod M, Magnussnon K, Lexner MO, Blomqvist S, Dahlén G, Twetman S. Oral microflora in infants delivered vaginally and by caesarean section. Int J Paediatr Dent. 2011;21:401–6.
Abiko Y. Passive immunization against dental caries and periodontal disease: development of recombinant and human monoclonal antibodies. Crit Rev Oral Biol Med. 2000;11:140–58.
Mathews MJ, Mathews EH, Mathews GE. Oral health and coronary heart disease. BMC Oral Health. 2016;16:122.
Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann Periodontol. 2003;8:38–53.
Demmer RT, Desvarieux M. Periodontal infections and cardiovascular disease. JADA. 2006;137:14–20.
Fong IW. Emerging relations between infectious diseases and coronary artery disease and atherosclerosis. CMAJ. 2000;163:49–56.
Huang X, Schulte RM, Burne RA, Nascimento MM. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015;49:165–76.
López-López A, Camelo-Castillo A, Ferrer MD, Simon-Soro Á, Mira A. Health-associated niche inhabitants as oral probiotics: the case of Streptococcus dentisani. Front Microbiol. 2017;8:379.
Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. Dental caries from a molecular microbiological perspective. Caries Res. 2013;47:89–102.
Abrahamsson TR, Jakobsson T, Björkstén B, Oldaeus G, Jenmalm MC. No effect of probiotics on respiratory allergies: a seven-year follow-up of a randomized controlled trial in infancy. Pediatr Allergy Immunol. 2013;24:556–61.
Abrahamsson TR, Jakobsson T, Böttcher MF, Fredrikson M, Jenmalm MC, Björkstén B, et al. Probiotics in prevention of IgE-associated eczema: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:1174–80.
Stensson M, Koch G, Coric S, Abrahamsson TR, Jenmalm MC, Birkhed D, et al. Oral administration of Lactobacillus reuteri during the first year of life reduces caries prevalence in the primary dentition at 9 years of age. Caries Res. 2014;48:111–7.
Alm A, Wendt LK, Koch G, Birkhed D. Prevalence of approximal caries in posterior teeth in 15-year-old Swedish teenagers in relation to their caries experience at 3 years of age. Caries Res. 2007;41:392–8.
Koch G. Effect of sodium fluoride in dentifrice and mouthwash on incidence of dental caries in schoolchildren. Odontol Rev. 1967;18:38–43.
Sipos R, Székely AJ, Palatinszky M, Révész S, Márialiget K, Nikolausz M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol Ecol. 2007;60:341–50.
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1.
Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4.
Edgar RC. UNCROSS: filtering of high-frequency cross-talk in 16S amplicon reads. bioRxiv. 2016. https://doi.org/10.1101/088666
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9.
Chen T, Yu W-H, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxf). 2010;2010:baq013.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42.
Oksanen J. Vegan: community ecology. R Package Version. 2018;2:4–6.
Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can Med Assoc J. 2013;185:385–94.
Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703.
Abrahamsson T, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm M. Reply: gut microbiota diversity and atopic disease: does breast-feeding play a role? J Allergy Clin Immunol. 2013;1:248–9.
Könönen E. Development of oral bacterial flora in young children. Ann Med. 2000;32:107–12.
Nelson-Filho P, Borba IG, Mesquita KSF de, Silva RAB, Queiroz AM de, Silva LAB, et al. Dynamics of microbial colonization of the oral cavity in newborns. Braz Dent J. 2013;24:415–9.
Lif Holgerson P, Harnevik L, Hernell O, Tanner ACR, Johansson I. Mode of birth delivery affects oral microbiota in infants. J Dent Res. 2011;90:1183–8.
Hyde MJ, Modi N. The long-term effects of birth by caesarean section: the case for a randomised controlled trial. Early Hum Dev. 2012;88:943–9.
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–5.
Amarasekare P, Possingham H. Patch dynamics and metapopulation theory: the case of successional species. J Theor Biol. 2001;209:333–44.
Luo AH, Yang DQ, Xin BC, Paster BJ, Qin J. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 2012;18:595–601.
Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6:e21313.
Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–51.
Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–904.
Jost T, Lacroix C, Braegger C, Chassard C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev. 2015;73:426–37.
Martin DH, Zozaya M, Lillis R, Miller J, Ferris MJ. The microbiota of the human genitourinary tract: trying to see the forest through the trees. Trans Am Clin Climatol Assoc. 2012;123:242–56.
Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and Bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr. 2014;59:78–88.
Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40.
Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197–205. https://doi.org/10.1038/nrgastro.2017.173
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32.
Mysak J, Podzimek S, Sommerova P, Lyuya-Mi Y, Bartova J, Janatova T, et al. Porphyromonas gingivalis: Major periodontopathic pathogen overview. J Immunol Res. 2014;2014:476068.
Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of ten healthy individuals. ISME J. 2010;4:962–74.
Holgerson PL, Vestman NR, Claesson R, Ohman C, Domellöf M, Tanner ACR, et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J Pediatr Gastroenterol Nutr. 2013;56:127–36.
Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17.
Alcaraz LD, Belda-Ferre P, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, et al. Identifying a healthy oral microbiome through metagenomics. Clin Microbiol Infect. 2012;18:54–57.
Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47–79.
Yasukawa T, Ohmori M, Sato S. 2010. The relationship between physiologic halitosis and periodontopathic bacteria of the tongue and gingival sulcus. Odontology 98:44–51.
Badet C, Thebaud NB. 2008. Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol. J. 2:38–48.
Bradshaw DJ, Marsh PD. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32:456–62.
Zaura E, Brandt BW, Teixeira de Mattos MJ, Buijs MJ, Caspers MPM, Rashid M-U, et al. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. 2015;6:e01693–15.
Simón-Soro Á, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47:591–600.
Mira A. Oral microbiome studies: potential diagnostic and therapeutic implications. Adv Dent Res. 2018;29(1):71–7. https://doi.org/10.1177/0022034517737024
Law V, Seow WK, Townsend G. Factors influencing oral colonization of mutans streptococci in young children. Aust Dent J. 2007;52:93–100.
Wan AKL, Seow WK, Purdie DM, Bird PS, Walsh LJ, Tudehope DI. Oral colonization of Streptococcus mutans in six-month-old predentate infants. J Dent Res. 2001a;80:2060–5.
Wan AKL, Seow WK, Walsh LJ, Bird P, Tudehope DI, Purdie DM. Association of Streptococcus mutans infection and oral developmental nodules in pre-dentate infants. J Dent Res. 2001b;80:1945–8.
Gruner D, Paris S, Schwendicke F. Probiotics for managing caries and periodontitis: systematic review and meta-analysis. J Dent. 2016;48:16–25.
Zbinden A, Bostanci N, Belibasakis GN. The novel species Streptococcus tigurinus and its association with oral infection. Virulence. 2015;6:177–82.
Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–8.
Burton JP, Wescombe PA, Moore CJ, Chilcott CN, Tagg JR. Safety assessment of the oral cavity probiotic Streptococcus salivarius K12. Appl Environ Microbiol. 2006;72:3050–3.
Martín V, Mañes-Lázaro R, Miguel Rodríguez J, Maldonado-Barragá A. Streptococcus lactarius sp. nov., isolated from breast milk of healthy women. Int J Syst Evol Microbiol. 2011;61:1048–52.
Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, Hardin JM. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018–23.
Coffey J, Shlossman M. Multiplex real‐time PCR detection and relative quantification of periodontal pathogens. Clin Exp Dent Res. 2016;2(3):185-92. https://doi.org/10.1002/cre2.37
Ho M-H, Lamont RJ, Xie H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci Rep. 2017;7:1413 https://doi.org/10.1038/s41598-017-01551-4
Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47:3902–6. https://doi.org/10.1128/JCM.00072-09
Mira A, Artacho A, Camelo-Castillo A, Garcia-Esteban S, Simon-Soro A. Salivary immune and metabolic marker analysis (SIMMA): a diagnostic test to predict caries risk. Diagnostics. 2017;7:38.
Young DA, Featherstone JDB. Implementing caries risk assessment and clinical interventions. Dent Clin North Am. 2010;54:495–505.
Kanasi E, Johansson I, Lu SC, Kressin NR, Nunn ME, Kent R, et al. Microbial risk markers for childhood caries in pediatricians’ offices. J Dent Res. 2010;89:378–83.
Simón-Soro Á, D’Auria G, Collado MC, Džunková M, Culshaw S, Mira A. Revealing microbial recognition by specific antibodies. BMC Microbiol. 2015;15:132.
Belda-Ferre P, Williamson J, Simón-Soro Á, Artacho A, Jensen ON, Mira A. The human oral metaproteome reveals potentialbiomarkers for caries disease. Proteomics. 2015;15:3497–507.
Van Best N, Hornef MW, Savelkoul PHM, Penders J. On the origin of species: factors shaping the establishment of infant’s gut microbiota. Birth Defects Res Part C Embryo Today Rev. 2015;105:240–51.
Acknowledgements
We would like to acknowledge the technical assistance performed by Ann-Marie Fornander and Camilla Janefjord. We would also like to thank Alba Boix-Amorós and Sandra Garcia-Esteban for their great assistance in the laboratory work.
Funding
A.M.: Spanish Ministry of Economy and Competitiveness (grant no. BIO2015-68711-R). M.S.: The Research Council for the South-East Sweden (grant no: 79001). M.C.J.: The Swedish Research Council (2016-01698); the Swedish Heart and Lung Foundation (20140321); the Medical Research Council of Southeast Sweden (FORSS-573471); the Cancer and Allergy Foundation. M.C.C.: European Research Council (ERC-starting grant 639226).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Dzidic, M., Collado, M.C., Abrahamsson, T. et al. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J 12, 2292–2306 (2018). https://doi.org/10.1038/s41396-018-0204-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-018-0204-z
This article is cited by
-
Full-length 16S rRNA gene sequencing by PacBio improves taxonomic resolution in human microbiome samples
BMC Genomics (2024)
-
Early life factors and oral microbial signatures define the risk of caries in a Swedish cohort of preschool children
Scientific Reports (2024)
-
Influence of maternal oral microbiome on newborn oral microbiome in healthy pregnancies
Italian Journal of Pediatrics (2023)
-
Development of the oral resistome during the first decade of life
Nature Communications (2023)
-
Mendelian randomization analyses reveal causal relationships between the human microbiome and longevity
Scientific Reports (2023)