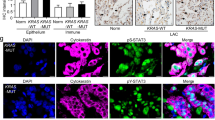
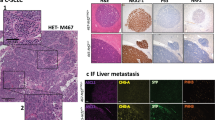
Abstract
Non-small cell lung cancer remains a highly lethal malignancy. Using the tamoxifen inducible Hnf1b:CreERT2 (H) transgenic mouse crossed to the LsL-KrasG12D (K) transgenic mouse, we recently discovered that an Hnf1b positive cell type in the lung is sensitive to adenoma formation when expressing a mutant KrasG12D allele. In these mice, we observe adenoma formation over a time frame of three to six months. To study specificity of the inducible Hnf1b:CreERT2 in the lung, we employed lineage tracing using an mTmG (G) reporter allele. This technique revealed recombined, GFP+ cells were predominantly SPC+. We further employed this technique in HKG mice to determine Hnf1b+ cells give rise to adenomas that express SPC and TTF1. Review of murine lung tissue confirmed a diagnosis of adenoma and early adenocarcinoma, a pathologic subtype of non-small cell lung cancer. Our expanded mouse model revealed loss of Mst1/2 promotes aggressive lung adenocarcinoma and large-scale proteomic analysis revealed upregulation of PKM2 in the lungs of mice with genetic deletion of Mst1/2. PKM2 is a known metabolic regulator in proliferating cells and cancer. Using a human lung adenocarcinoma cell line, we show pharmacologic inhibition of Mst1/2 increases the abundance of PKM2, indicating genetic loss or pharmacologic inhibition of Mst1/2 directly modulates the abundance of PKM2. In conclusion, here we report a novel model of non-small cell lung cancer driven by a mutation in Kras and deletion of Mst1/2 kinases. Tumor development is restricted to a subset of alveolar type II cells expressing Hnf1b. Our data show loss of Mst1/2 regulates levels of a potent metabolic regulator, PKM2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203 e13.
Sheridan C, Downward J. Overview of KRAS-driven genetically engineered mouse models of non-small cell lung cancer. Curr Protoc Pharm. 2015;70:14.35.1–16.
Vasan N, Boyer JL, Herbst RS. A RAS renaissance: emerging targeted therapies for KRAS-mutated non-small cell lung cancer. Clin Cancer Res. 2014;20:3921–30.
Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696.
Zhou JX, Yang H, Deng Q, Gu X, He P, Lin Y, et al. Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann Oncol. 2013;24:1319–25.
Abdulkareem FB, Sanni LA, Richman SD, Chambers P, Hemmings G, Grabsch H, et al. KRAS and BRAF mutations in Nigerian colorectal cancers. West Afr J Med 2012;31:198–203.
Xu C, Liu YL, Huang J, He DM, Hou YY, Ji Y, et al. [Detection of KRAS gene mutation and its clinical significance in colorectal adenocarcinoma]. Zhonghua Bing Li Xue Za Zhi. 2012;41:667–70.
Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67.
Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Investig. 2012;122:639–53.
Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–85.
Kimmelman AC. Metabolic Dependencies in RAS-Driven Cancers. Clin Cancer Res. 2015;21:1828–34.
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Disco. 2014;13:828–51.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Johansson L. Histopathologic classification of lung cancer: relevance of cytokeratin and TTF-1 immunophenotyping. Ann Diagn Pathol. 2004;8:259–67.
Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–8.
Xu CM, Liu WW, Liu CJ, Wen C, Lu HF, Wan FS. Mst1 overexpression inhibited the growth of human non-small cell lung cancer in vitro and in vivo. Cancer Gene Ther. 2013;20:453–60.
Yeung B, Yu J, Yang X. Roles of the Hippo pathway in lung development and tumorigenesis. Int J Cancer. 2016;138:533–9.
Ehmer U, Sage J. Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway. Mol Cancer Res. 2016;14:127–40.
Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803.
Santinon G, Pocaterra A, Dupont S. Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways. Trends Cell Biol. 2016;26:289–99.
Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–9.
Chen HY, Yu SL, Ho BC, Su KY, Hsu YC, Chang CS, et al. R331W missense mutation of oncogene YAP1 is a germline risk allele for lung adenocarcinoma with medical actionability. J Clin Oncol. 2015;33:2303–10.
Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–97.
Teoh SL, Das S. The emerging role of the hippo pathway in lung cancers: clinical implications. Curr Drug Targets. 2017;18:1880–92.
Ye S, Eisinger-Mathason TS. Targeting the hippo pathway: clinical implications and therapeutics. Pharm Res. 2016;103:270–8.
Guo L, Teng L. YAP/TAZ for cancer therapy: opportunities and challenges (review). Int J Oncol. 2015;46:1444–52.
Ma Y, Yang Y, Wang F, Wei Q, Qin H. Hippo-YAP signaling pathway: a new paradigm for cancer therapy. Int J Cancer. 2015;137:2275–86.
Pan D. The Hippo signaling pathway in development and cancer. Developmental Cell. 2010;19:491–505.
Schneider J, Neu K, Velcovsky HG, Morr H, Eigenbrodt E. Tumor M2-pyruvate kinase in the follow-up of inoperable lung cancer patients: a pilot study. Cancer Lett. 2003;193:91–8.
Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci USA. 2014;111:4952–7.
De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development. 2015;142:871–82.
Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–85.
Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629.
Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–38.
Jeong H, Kim S, Hong BJ, Lee CJ, Kim YE, Bok S, et al. Tumor-associated macrophages enhance tumor hypoxia and aerobic glycolysis. Cancer Res. 2019;79:795–806.
Fritz JM, Tennis MA, Orlicky DJ, Lin H, Ju C, Redente EF, et al. Depletion of tumor-associated macrophages slows the growth of chemically induced mouse lung adenocarcinomas. Front Immunol. 2014;5:587.
Ju R, Wu D, Guo L, Li J, Ye C, Zhang D. Inhibition of pro-inflammatory cytokines in tumour associated macrophages is a potential anti-cancer mechanism of carboxyamidotriazole. Eur J Cancer. 2012;48:1085–95.
Wang R, Zhang J, Chen S, Lu M, Luo X, Yao S, et al. Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer. 2011;74:188–96.
Cooper WN, Hesson LB, Matallanas D, Dallol A, von Kriegsheim A, Ward R, et al. RASSF2 associates with and stabilizes the proapoptotic kinase MST2. Oncogene. 2009;28:2988–98.
Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, et al. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007;26:4523–34.
Ciuffreda L, Incani UC, Steelman LS, Abrams SL, Falcone I, Curatolo AD, et al. Signaling intermediates (MAPK and PI3K) as therapeutic targets in NSCLC. Curr Pharm Des. 2014;20:3944–57.
Creighton CJ, Huang S. Reverse phase protein arrays in signaling pathways: a data integration perspective. Drug Des Devel Ther. 2015;9:3519–27.
Du D, Ma W, Yates MS, Chen T, Lu KH, Lu Y, et al. Predicting high-risk endometrioid carcinomas using proteins. Oncotarget. 2018;9:19704–15.
Gomez DR, Byers LA, Nilsson M, Diao L, Wang J, Li L, et al. Integrative proteomic and transcriptomic analysis provides evidence for TrkB (NTRK2) as a therapeutic target in combination with tyrosine kinase inhibitors for non-small cell lung cancer. Oncotarget. 2018;9:14268–84.
Clarke CN, Lee MS, Wei W, Manyam G, Jiang ZQ, Lu Y, et al. Proteomic features of colorectal cancer identify tumor subtypes independent of oncogenic mutations and independently predict relapse-free survival. Ann Surg Oncol. 2017;24:4051–8.
Zhang L, Wei Q, Mao L, Liu W, Mills GB, Coombes K. Serial dilution curve: a new method for analysis of reverse phase protein array data. Bioinformatics. 2009;25:650–4.
Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, et al. PKM2 and cancer: the function of PKM2 beyond glycolysis. Oncol Lett. 2016;11:1980–6.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63.
Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73.
Liu VM, Vander Heiden MG. The role of pyruvate kinase M2 in cancer metabolism. Brain Pathol. 2015;25:781–3.
Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12:3154–8.
Rizvi S, Yamada D, Hirsova P, Bronk SF, Werneburg NW, Krishnan A, et al. A hippo and fibroblast growth factor receptor autocrine pathway in cholangiocarcinoma. J Biol Chem. 2016;291:8031–47.
Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18:5318–24.
Miller MS. Transplacental lung carcinogenesis: a pharmacogenetic mouse model for the modulatory role of cytochrome P450 1A1 on lung cancer initiation. Chem Res Toxicol. 1994;7:471–81.
Malkinson AM. The genetic basis of susceptibility to lung tumors in mice. Toxicology. 1989;54:241–71.
Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–64.
Kwak I, Tsai SY, DeMayo FJ. Genetically engineered mouse models for lung cancer. Annu Rev Physiol. 2004;66:647–63.
Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22.
Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34.
Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–10.
Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, et al. The hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–22.
Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, et al. Expression of yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–9.
Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci USA. 2011;108:E1312–20.
Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–71.
Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–42.
Kim HB, Myung SJ. Clinical implications of the Hippo-YAP pathway in multiple cancer contexts. BMB Rep. 2018;51:119–25.
Li RZ, Fan XX, Shi DF, Zhu GY, Wang YW, Luo LX, et al. Identification of a new pyruvate kinase M2 isoform (PKM2) activator for the treatment of non-small-cell lung cancer (NSCLC). Chem Biol Drug Des. 2018;92:1851–8.
Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57:95–107.
Li H, Xu H, Xing R, Pan Y, Li W, Cui J, et al. Pyruvate kinase M2 contributes to cell growth in gastric cancer via aerobic glycolysis. Pathol Res Pr. 2019;215:152409.
Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23:1362–8.
Polireddy K, Singh K, Pruski M, Jones NC, Manisundaram NV, Ponnela P, et al. Mutant p53. Cancer Lett. 2019;453:122–30.
Acknowledgements
We would like to thank Bingqian Hu and Junlan Zhang for assistance with lung perfusions. We also thank Naveen Manisundaram for intellectual insights and discussions. JMB is supported by the AACR/Pancreatic Cancer Action Network Pathway to Leadership Award. NCJ is supported by NIH T35 DK007676-22. HKE is supported by National Institute of Health Grants R01 DK097075, POI-HL114457, R01-HL109233, R01-DK109574, R01-HL119837, and R01-HL133900.
Author information
Authors and Affiliations
Contributions
KS and MAP were involved in data acquisition and manuscript writing. KP, NCJ, QC, JY all assisted in data acquisition. WAD, HK-Q, and HKE intellectually contributed to the manuscript and assisted in writing. MY was involved in data acquisition. CM was involved in data analysis and acquisition. FM and CJ contributed intellectually and contributed to manuscript writing. HY contributed intellectually and was involved in data analysis and acquisition. JMB oversaw all aspects of the project including data acquisition, intellectual contribution, and manuscript writing and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Singh, K., Pruski, M.A., Polireddy, K. et al. Mst1/2 kinases restrain transformation in a novel transgenic model of Ras driven non-small cell lung cancer. Oncogene 39, 1152–1164 (2020). https://doi.org/10.1038/s41388-019-1031-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1031-z
This article is cited by
-
Kras mutation rate precisely orchestrates ductal derived pancreatic intraepithelial neoplasia and pancreatic cancer
Laboratory Investigation (2021)