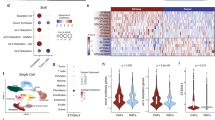
Abstract
Integrin beta 4 (ITGB4) overexpression in cancer cells contributes to cancer progression. However, the role of stromal ITGB4 expression in cancer progression remains poorly understood, despite stromal ITGB4 overexpression in malignant cancers. In our study, ITGB4-overexpressing triple negative breast cancer (TNBC) cells provided cancer-associated fibroblasts (CAFs) with ITGB4 proteins via exosomes, which induced BNIP3L-dependent mitophagy and lactate production in CAFs. In coculture assays, the ITGB4-induced mitophagy and glycolysis were suppressed in CAFs by knocking down ITGB4 or inhibiting exosome generation in MDA-MB-231, or blocking c-Jun or AMPK phosphorylation in CAFs. ITGB4-overexpressing CAF-conditioned medium promoted the proliferation, epithelial-to-mesenchymal transition, and invasion of breast cancer cells. In a co-transplant mouse model, MDA-MB-231 made a bigger tumor mass with CAFs than ITGB4 knockdown MDA-MB-231. Herein, we presented how TNBC-derived ITGB4 protein triggers glycolysis in CAFs via BNIP3L-dependent mitophagy and suggested the possibility that ITGB4-induced mitophagy could be targeted as a cancer therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ratnikov BI, Scott DA, Osterman AL, Smith JW, Ronai ZA. Metabolic rewiring in melanoma. Oncogene. 2017;36:147–57.
Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001.
Martins D, Beca FF, Sousa B, Baltazar F, Paredes J, Schmitt F. Loss of caveolin-1 and gain of MCT4 expression in the tumor stroma: key events in the progression from an in situ to an invasive breast carcinoma. Cell Cycle. 2013;12:2684–90.
Witkiewicz AK, Dasgupta A, Nguyen KH, Liu C, Kovatich AJ, Schwartz GF, et al. Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol Ther. 2009;8:1071–9.
Wu KN, Queenan M, Brody JR, Potoczek M, Sotgia F, Lisanti MP, et al. Loss of stromal caveolin-1 expression in malignant melanoma metastases predicts poor survival. Cell Cycle. 2011;10:4250–5.
Fu Y, Liu S, Yin S, Niu W, Xiong W, Tan M, et al. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget. 2017;8:57813–25.
Martinez-Outschoorn UE, Lin Z, Trimmer C, Flomenberg N, Wang C, Pavlides S, et al. Cancer cells metabolically “fertilize” the tumor microenvironment with hydrogen peroxide, driving the Warburg effect: implications for PET imaging of human tumors. Cell Cycle. 2011;10:2504–20.
Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93.
Kim BG, Gao MQ, Choi YP, Kang S, Park HR, Kang KS, et al. Invasive breast cancer induces laminin-332 upregulation and integrin beta4 neoexpression in myofibroblasts to confer an anoikis-resistant phenotype during tissue remodeling. Breast Cancer Res. 2012;14:R88.
Yang L, Hou Y, Yuan J, Tang S, Zhang H, Zhu Q, et al. Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget. 2015;6:25755–69.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9.
Peterkofsky B, Prather W. Correlation between the rates of aerobic glycolysis and glucose transport, unrelated to neoplastic transformation, in a series of BALB 3T3-derived cell lines. Cancer Res. 1982;42:1809–16.
Maycotte P, Marin-Hernandez A, Goyri-Aguirre M, Anaya-Ruiz M, Reyes-Leyva J, Cortes-Hernandez P. Mitochondrial dynamics and cancer. Tumour Biol. 2017;39:1010428317698391.
Esteban-Martinez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Marino G, Seco E, et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017;36:1688–706.
Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111:1208–21.
Quelo I, Gauthier C, Hannigan GE, Dedhar S, St-Arnaud R. Integrin-linked kinase regulates the nuclear entry of the c-Jun coactivator alpha-NAC and its coactivation potency. J Biol Chem. 2004;279:43893–9.
Ke H, Harris R, Coloff JL, Jin JY, Leshin B, Miliani de Marval P, et al. The c-Jun NH2-terminal kinase 2 plays a dominant role in human epidermal neoplasia. Cancer Res. 2010;70:3080–8.
Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51.
Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–8.
Roach PJ. AMPK -> ULK1 -> autophagy. Mol Cell Biol. 2011;31:3082–4.
Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–28.
Liu M, Quek LE, Sultani G, Turner N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016;4:19.
Kim BG, An HJ, Kang S, Choi YP, Gao MQ, Park H, et al. Laminin-332-rich tumor microenvironment for tumor invasion in the interface zone of breast cancer. Am J Pathol. 2011;178:373–81.
Kim BG, Gao MQ, Kang S, Choi YP, Lee JH, Kim JE, et al. Mechanical compression induces VEGFA overexpression in breast cancer via DNMT3A-dependent miR-9 downregulation. Cell Death Dis. 2017;8:e2646.
Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–67.
Jezek J, Cooper KF, Strich R. Reactive oxygen species and mitochondrial dynamics: the Yin and Yang of mitochondrial dysfunction and cancer progression. Antioxidants. 2018;7:1–24.
Renschler MF. The emerging role of reactive oxygen species in cancer therapy. Eur J Cancer. 2004;40:1934–40.
Takahashi A, Ohtani N, Yamakoshi K, Iida S, Tahara H, Nakayama K, et al. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat Cell Biol. 2006;8:1291–7.
Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823:2297–310.
Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–96.
Li XL, Liu L, Li DD, He YP, Guo LH, Sun LP, et al. Integrin beta4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Sci Rep. 2017;7:1–12.
McKeown SR. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radio. 2014;87:20130676.
Pavlides S, Vera I, Gandara R, Sneddon S, Pestell RG, Mercier I, et al. Warburg meets autophagy: cancer-associated fibroblasts accelerate tumor growth and metastasis via oxidative stress, mitophagy, and aerobic glycolysis. Antioxid Redox Signal. 2012;16:1264–84.
Xiao B, Goh JY, Xiao L, Xian H, Lim KL, Liou YC. Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J Biol Chem. 2017;292:16697–708.
Cicenas J, Zalyte E, Rimkus A, Dapkus D, Noreika R, Urbonavicius S. JNK, p38, ERK, and SGK1 inhibitors in cancer. Cancers. 2017;10:1–12.
Liu SY, Ge D, Chen LN, Zhao J, Su L, Zhang SL, et al. A small molecule induces integrin beta4 nuclear translocation and apoptosis selectively in cancer cells with high expression of integrin beta4. Oncotarget. 2016;7:16282–96.
Kosgodage US, Mould R, Henley AB, Nunn AV, Guy GW, Thomas EL, et al. Cannabidiol (CBD) Is a novel inhibitor for exosome and microvesicle (EMV) release in cancer. Front Pharm. 2018;9:889.
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4.
Vallejo AN, Pogulis RJ, Pease LR. PCR mutagenesis by overlap extension and gene SOE. CSH Protoc. 2008;2008:pdbprot4861.
Acknowledgements
This study was supported by the Mid-Career Researcher Program (no. 2019R1A2B5B01069934; NHC) and Challenges in Creative Research (no. 2019R1I1A1A01060549; BGK) through a National Research Foundation of Korea grant, and The Health Fellowship Foundation grants funded by Yuhan Corporation. The authors thank Medical Illustration & Design, a part of the Medical Research Support Services of Yonsei University College of Medicine, for all artistic support related to this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sung, J.S., Kang, C.W., Kang, S. et al. ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 39, 664–676 (2020). https://doi.org/10.1038/s41388-019-1014-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1014-0
This article is cited by
-
Machine learning reveals diverse cell death patterns in lung adenocarcinoma prognosis and therapy
npj Precision Oncology (2024)
-
ITGB4 upregulation is associated with progression of lower grade glioma
Scientific Reports (2024)
-
Exosome-transmitted FOSL1 from cancer-associated fibroblasts drives colorectal cancer stemness and chemo-resistance through transcriptionally activating ITGB4
Molecular and Cellular Biochemistry (2024)
-
Integrin β4 Regulates Cell Migration of Lung Adenocarcinoma Through FAK Signaling
Molecular Biotechnology (2024)
-
Integrin signaling in cancer: bidirectional mechanisms and therapeutic opportunities
Cell Communication and Signaling (2023)