Abstract
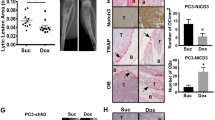
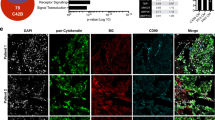
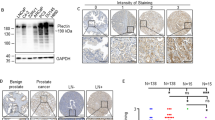
We previously showed that KLF4, a gene highly expressed in murine prostate stem cells, blocks the progression of indolent intraepithelial prostatic lesions into aggressive and rapidly growing tumors. Here, we show that the anti-tumorigenic effect of KLF4 extends to PC3 human prostate cancer cells growing in the bone. We compared KLF4 null cells with cells transduced with a DOX-inducible KLF4 expression system, and find KLF4 function inhibits PC3 growth in monolayer and soft agar cultures. Furthermore, KLF4 null cells proliferate rapidly, forming large, invasive, and osteolytic tumors when injected into mouse femurs, whereas KLF4 re-expression immediately after their intra-femoral inoculation blocks tumor development and preserves a normal bone architecture. KLF4 re-expression in established KLF4 null bone tumors inhibits their osteolytic effects, preventing bone fractures and inducing an osteogenic response with new bone formation. In addition to these profound biological changes, KLF4 also induces a transcriptional shift from an osteolytic program in KLF4 null cells to an osteogenic program. Importantly, bioinformatic analysis shows that genes regulated by KLF4 overlap significantly with those expressed in metastatic prostate cancer patients and in three individual cohorts with bone metastases, strengthening the clinical relevance of the findings in our xenograft model.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30.
Vela I, Gregory L, Gardiner EM, Clements JA, Nicol DL. Bone and prostate cancer cell interactions in metastatic prostate cancer. BJU Int 2007;99:735–42.
Crockett JC, Rogers MJ, Coxon FP, Hocking LJ, Helfrich MH. Bone remodelling at a glance. J Cell Sci 2011;124:991–8.
Park SH, Keller ET, Shiozawa Y. Bone marrow microenvironment as a regulator and therapeutic target for prostate cancer bone metastasis. Calcif Tissue Int 2018;102:152–62.
Sims NA. Cell-specific paracrine actions of IL-6 family cytokines from bone, marrow and muscle that control bone formation and resorption. Int J Biochem Cell Biol 2016;79:14–23.
Gooding S, Edwards CM. New approaches to targeting the bone marrow microenvironment in multiple myeloma. Curr Opin Pharmacol 2016;28:43–9.
Nandana S, Tripathi M, Duan P, Chu C-Y, Mishra R, Liu C, et al. Bone metastasis of prostate cancer can be therapeutically targeted at the TBX2-WNT signaling axis. Cancer Res 2017;77:1331–44.
Wu JB, Yin L, Shi C, Li Q, Duan P, Huang J-M, et al. MAOA-dependent activation of Shh-IL6-RANKL signaling network promotes prostate cancer metastasis by engaging tumor-stromal cell interactions. Cancer Cell 2017;31:368–82.
Body J-J, Casimiro S, Costa L. Targeting bone metastases in prostate cancer: improving clinical outcome. Nat Rev Urol 2015;12:340–56.
Xiong X, Schober M, Tassone E, Khodadadi-Jamayran A, Sastre Perona A, Zhou H, et al. KLF4, A gene regulating prostate stem cell homeostasis, is a barrier to malignant progression and predictor of good prognosis in prostate cancer. Cell Rep 2018;25:3006–20.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–76.
Liu YN, Abou-Kheir W, Yin JJ, Fang L, Hynes P, Casey O, et al. Critical and reciprocal regulation of KLF4 and SLUG in transforming growth factor β-initiated prostate cancer epithelial-mesenchymal transition. Mol Cell Biol 2012;32:941–53.
Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE, et al. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res 2010;70:10182–91.
Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer 2005;6:11–23.
Tetreault M-P, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer 2013;13:701–13.
Ghaleb AM, Yang VW. Krüppel-like factor 4 (KLF4): what we currently know. Gene 2017;611:27–37.
Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol 1979;17:16–23.
Siu M-K, Suau F, Chen W-Y, Tsai Y-C, Tsai H-Y, Yeh H-L, et al. KLF4 functions as an activator of the androgen receptor through reciprocal feedback. Oncogenesis 2016;5:e282.
Sanchez-Sweatman OH, Orr FW, Singh G. Human metastatic prostate PC3 cell lines degrade bone using matrix metalloproteinases. Invasion Metastasis 1998;18:297–305.
Armstrong AP, Miller RE, Jones JC, Zhang J, Keller ET, Dougall WC. RANKL acts directly on RANK-expressing prostate tumor cells and mediates migration and expression of tumor metastasis genes. Prostate 2007;68:92–104.
Kudo O, Fujikawa Y, Itonaga I, Sabokbar A, Torisu T, Athanasou NA. Proinflammatory cytokine (TNFα/IL-1α) induction of human osteoclast formation. J Pathol 2002;198:220–7.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22.
Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64.
Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78.
Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate. Cancer Cell 2018;174:758–769.e9.
Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. May
Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Krüppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol 2003;326:665–77.
Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005;7:1074–82.
Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 2004;23:395–402.
Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, et al. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res 2005;65:2746–54.
Sastre-Perona A, Hoang-Phou S, Leitner MC, Okuniewska M, Meehan S, Schober M. De novo PITX1 expression controls bi-stable transcriptional circuits to govern self-renewal and differentiation in squamous cell carcinoma. Cell Stem Cell 2019;24:390–404.
Fradet A, Sorel H, Depalle B, Serre CM, Farlay D, Turtoi A, et al. A new murine model of osteoblastic/osteolytic lesions from human androgen-resistant prostate cancer. PLoS ONE 2013;8:e75092.
Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J Clin Invest 2008;118:2697–710.
LeRoy BE, Thudi NK, Nadella MVP, Toribio RE, Tannehill-Gregg SH, van Bokhoven A, et al. New bone formation and osteolysis by a metastatic, highly invasive canine prostate carcinoma xenograft. Prostate 2006;66:1213–22.
Yoshida T, Yamashita M, Hayashi M. Kruppel-like factor 4 contributes to high phosphate-induced phenotypic switching of vascular smooth muscle cells into osteogenic cells. J Biol Chem 2012;287:25706–14.
Barba M, Pirozzi F, Saulnier N, Vitali T, Natale MT, Logroscino G, et al. Lim mineralization protein 3 induces the osteogenic differentiation of human amniotic fluid stromal cells through kruppel-like factor-4 downregulation and further bone-specific gene expression. J Biomed Biotechnol 2012;2012:1–11.
Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003;423:337–42.
Sottnik JL, Keller ET. Understanding and targeting osteoclastic activity in prostate cancer bone metastases. Curr Mol Med. 2013;13:626–39.
Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PVN, Komm BS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem 2005;280:33132–40.
Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol 2007;26:508–23.
Tsuji T, Kondo E, Yasoda A, Inamoto M, Kiyosu C, Nakao K, et al. Hypomorphic mutation in mouse Nppc gene causes retarded bone growth due to impaired endochondral ossification. Biochem Biophys Res Commun 2008;376:186–90.
Rucci N, Rufo A, Alamanou M, Capulli M, Del Fattore A, Åhrman E, et al. The glycosaminoglycan-binding domain of PRELP acts as a cell type-specific NF-κB inhibitor that impairs osteoclastogenesis. J Cell Biol 2009;187:669–83.
Uchimura T, Hollander JM, Nakamura DS, Liu Z, Rosen CJ, Georgakoudi I, et al. An essential role for IGF2 in cartilage development and glucose metabolism during postnatal long bone growth. Development 2017;144:3533–46.
Morcos MW, Al-Jallad H, Li J, Farquharson C, Millán JL, Hamdy RC, et al. PHOSPHO1 is essential for normal bone fracture healing. Bone Jt Res 2018;7:397–405.
Osta B, Benedetti G, Miossec P. Classical and paradoxical effects of TNF-α on bone homeostasis. Front Immunol 2014;5:48.
Lee H-L, Yi T, Woo KM, Ryoo H-M, Kim G-S, Baek J-H. Msx2 mediates the inhibitory action of TNF-α on osteoblast differentiation. Exp Mol Med 2010;42:437–9.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308.
Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 2008;3:346–53.
Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp 2014;92:1–6.
Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Min Res 2010;25:1468–86.
Leucht P, Kim J-B, Wazen R, Currey JA, Nanci A, Brunski JB, et al. Effect of mechanical stimuli on skeletal regeneration around implants. Bone 2007;40:919–30.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2012;29:15–21.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2008;37:1–13.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4.
Acknowledgements
We thank Dr. E. Hernando-Monge (NYU School of Medicine) for the GFP-luciferase plasmid (BLIV513PA-1) and Drs. S. Logan and M. Garabedian (NYU School of Medicine) for helpful discussions. This study was supported by the NIH (R01CA132641 to E.L.W., R01AG056169 and K08AR069099 to P.L., R01CA181111 to M.S., T32CA009161 and T32AR064184 to A.S-P.), the American Cancer Society (RSG-16-033-01-DDC to M.S.), the Department of Urology and the Kimmel Center for Stem Cell Biology. Core funding (Applied Bioinformatics Laboratories, Genome Technology Center, High Throughput Biology Laboratory, Experimental Pathology Research Laboratory, Micro-CT) is partially supported by the Perlmutter Cancer Center (P30CA016087), NYSTEM (contract C026719), and NIH (S10 OD010751).
Author information
Authors and Affiliations
Contributions
E.T., P.L., M.S., and E.L.W. conceived and designed the study. E.T., V.B-C., X.X., and A.M.J. performed the experiments and analyzed the data. M.S., A.S-P., and A.K-J. performed the bioinformatics analysis. J.M. and P.L. interpreted histological data. L.B. provided the BFP sequence of the KLF4-expressing vector and helped with the cloning strategy. D.J.K. acquired the images of soft agar cultures and analyzed the data. E.T., L.O., M.S., and E.L.W. wrote the manuscript. All authors revised and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tassone, E., Bradaschia-Correa, V., Xiong, X. et al. KLF4 as a rheostat of osteolysis and osteogenesis in prostate tumors in the bone. Oncogene 38, 5766–5777 (2019). https://doi.org/10.1038/s41388-019-0841-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0841-3