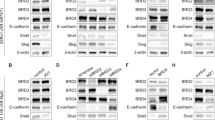
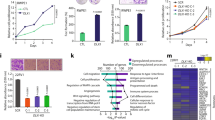
Abstract
BET bromodomain inhibitors block prostate cancer cell growth at least in part through c-Myc and androgen receptor (AR) suppression. However, little is known about other transcriptional regulators whose suppression contributes to BET bromodomain inhibitor anti-tumor activity. Moreover, the anti-tumor activity of BET bromodomain inhibition in AR-independent castration-resistant prostate cancers (CRPC), whose frequency is increasing, is also unknown. Herein, we demonstrate that BET bromodomain inhibition blocks growth of a diverse set of CRPC cell models, including those that are AR-independent or in which c-Myc is not suppressed. To identify transcriptional regulators whose suppression accounts for these effects, we treated multiple CRPC cell lines with the BET bromodomain inhibitor JQ1 and then performed RNA-sequencing followed by Master Regulator computational analysis. This approach identified several previously unappreciated transcriptional regulators that are highly expressed in CRPC and whose suppression, via both transcriptional or post-translational mechanisms, contributes to the anti-tumor activity of BET bromodomain inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
American Cancer Society. Cancer Facts and Figures 2019. (2019)
Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–95.
Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol. 2018;36:2492–503.
Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular cell. 2005;19:523–34.
Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82.
Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–97.
Coleman DJ, Hook K Van, King CJ, Schwartzman J, Lisac R, Urrutia J. Cellular androgen content influences enzalutamide agonism of F877L mutant androgen receptor. Oncotarget. 2016;7:40690–40703.
Gao L, Schwartzman J, Gibbs A, Lisac R, Kleinschmidt R, Wilmot B, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS ONE. 2013;8:e63563.
Wyce A, Degenhardt Y, Bai Y, Le B, Korenchuk S, Crouthame MC, et al. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget. 2013;4:2419–29.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17.
Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clinical Cancer Research. 2014;20:912–25.
Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci. 2011;108:16669–74.
Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:377.
MJ Alvarez. viper: Virtual Inference of Protein-activity by Enriched Regulon analysis. Bioconductor. 2013. Vol. R package, p. Software.
Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–51.
Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–47.
Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73.
MJ Alvarez, F Giorgi, and A Califano. Using viper, a package for virtual inference of protein-activity by enriched regulon analysis. Bioconductor. 2014:1-14
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–25.
Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016;13:521–7.
Kuruma H, Matsumoto H, Shiota M, Bishop J, Lamoureux F, Thomas C, et al. A novel antiandrogen, Compound 30, suppresses castration-resistant and MDV3100-resistant prostate cancer growth in vitro and in vivo. Mol Cancer Ther. 2013;12:567–76.
Bishop JL, Sio A, Angeles A, Roberts ME, Azad AA, Chi KN, et al. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. Oncotarget. 2015;6:234–42.
Culig Z, Hoffmann J, Erdel M, Eder IE, Hobisch A, Hittmair A, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer. 1999;81:242–51.
Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62.
Snoek R, Cheng H, Margiotti K, Wafa LA, Wong CA, Wong EC, et al. In vivo knockdown of the androgen receptor results in growth inhibition and regression of well-established, castration-resistant prostate tumors. Clin Cancer Res. 2009;15:39–47.
Coleman DJ, Gao L, Schwartzman J, Korkola JE, Sampson D, Derrick DS, et al. Maintenance of MYC expression promotes de novo resistance to BET bromodomain inhibition in castration-resistant prostate cancer. Sci Rep. 2019;9:3823.
Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43.
Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406.
Liu X, Erikson RL. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci USA. 2003;100:5789–94.
Valensin S, Ghiron C, Lamanna C, Kremer A, Rossi M, Ferruzzi P, et al. KIF11 inhibition for glioblastoma treatment: reason to hope or a struggle with the brain?. BMC Cancer. 2009;9:196.
Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem cells. 2007;25:3121–32.
Ibarra A, Schwob E, Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA. 2008;105:8956–61.
Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–8.
Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23.
Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JM, Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426–36.
Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52:25–36.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–89.e6
Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–31.
Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, et al. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:6154–63.
Zhang JJ, Zhao Y, Chait BT, Lathem WW, Ritzi M, Knippers R, et al. Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J. 1998;17:6963–71.
Rathert P, Roth M, Neumann T, Muerdter F, Roe J-S, Muhar M, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–7.
Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–42.
Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM. et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer Nature. 2016;529:413–417.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and 747 correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9.
Acknowledgements
This research was supported by: National Institutes of Health (NIH)/National Cancer Institute (NCI) R01 award CA178610; the Pacific Northwest Prostate Cancer SPORE/NCI (P50 CA097186); Cancer Center Support Grant (CCSG) (P30 CA069533); the Oregon Clinical and Translational Research Institute (OCTRI) (UL1TR000128) from the National Center for Advancing Translational Sciences (NCATS), a component of the NIH; Department of Defense Synergistic Idea Award (W81XWH-13-1-0420); Department of Defense Impact Awards (W81XWH-16-1-0597 and W81XWH-16-1-1601). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or DOD. Other support includes: Wayne D. Kuni and Joan E. Kuni Foundation, Prospect Creek Foundation, a Hope Foundation Award, and a Stand Up to Cancer—Prostate Cancer Foundation—Prostate Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0409); this research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (7465sc). We thank Dr. James Bradner at Dana-Farber Cancer Institute for providing the JQ1 compound used in in vivo studies.
Author information
Authors and Affiliations
Contributions
DJC, LG, JS, LMH, and JJA designed the research; DJC, LG, JS, JU, AS, and JT performed the experiments; DJC, LG, CJK, JS, AB, JB, KEC, DSD, DS, ZX, LMH, and JJA analyzed the data, DJC, LG, CJK, JS, AB, LMH, and JJA provided critical analysis of the manuscript; DJC and JJA wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
JJA has performed consulting or held an advisory role with Astellas Pharma, Bayer, and Janssen Biotech, Inc. OHSU has received institutional research funding from Aragon Pharmaceuticals Inc., Astellas Pharma, Norvartis, Zenith Epigenetics Ltd, and Gilead Sciences Inc. These interests had no role in the design or conduct of this study. The remaining authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Coleman, D.J., Gao, L., King, C.J. et al. BET bromodomain inhibition blocks the function of a critical AR-independent master regulator network in lethal prostate cancer. Oncogene 38, 5658–5669 (2019). https://doi.org/10.1038/s41388-019-0815-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0815-5
This article is cited by
-
A census of pathway maps in cancer systems biology
Nature Reviews Cancer (2020)