Abstract
Recently, nuclear poly-ADP-ribosylation had aroused research interest in epigenetics, but little attempt to explore functions of mono-ADP-ribosylation of histone, the major formation of histone ADP-ribosylated modification. We have previously reported a novel mono-ADP-ribosylation of H3R117, which promoted proliferation of LoVo cells. Here we showed that mono-ADP-ribosylated H3R117 of LoVo cells depressed demethylation of tumor suppressor TFPI2 promoter by suppressing TET1 expression and adjusting H3K9me3 enrichment of TFPI2 promoter to attenuate affinity of TET1, besides, since high H3K27me3 level was associated with hypermethylation, mono-ADP-ribosylated-H3R117-depended-H3K27me3 of TFPI2 promoter may contribute to hypermethylation of TFPI2. However, H3R117A mutation increased poly-ADP-ribosylated modification of TET1 promoter not TFPI2 promoter, which resulted in boosting transcription and expression of TET1 by altering DNA methylated modification, chromatin accessibility, and histone-methylated modification of TET1 promoter, while knockout TET1 of H3R117A LoVo cells directly led to hypermethylation of TFPI2 promoter and depression of TFPI2 secretion as well as enhanced proliferation, suggested that TET1 played a key role in demethylation of TFPI2, production of TFPI2, and cell proliferation. Bioinformatics analyses reveal prevalent hypermethylation of TFPI2 was an early event in tumorigenesis of colorectal caner, and expression of TET1 and TFPI2 was positive correlation in colorectal cancer and normal tissue. These data suggested that mono-ADP-ribosylation of H3R117 upregulated methylation of TFPI2 by impact TET1, since hypermethyaltion of TFPI2 was an early event in tumorigenesis, selectively target mono-ADP-ribosylation of H3R117 deficiency could be a feasible way to block tumorigenesis of colorectal cancer.
Similar content being viewed by others
Introduction
Tissue factor pathway inhibitor-2 (TFPI2), a Kunitz-type serine proteinase inhibitor, can inhibit a variety of serine proteases including factor VIIa/tissue factor, factor Xa, plasmin, trypsin, chymotryspin, and plasma kallikrein. During the past decade studies, TFPI2 has been identified as a tumor suppressor gene (TSG) in several types of cancer, including colorectal cancer (CRC) [1,2,3]. TFPI2 methylation frequently existed in CRC patients’ sera [4] and stool [5]. Moreover, hypermethylated TFPI2 was associated with recurrence and early stage of CRC [6], besides, TFPI2 was significant in CRC patients’ sera with large, poorly differentiated carcinoma, deep invasion, lymph node metastasis, or distant metastasis [4]. Additionally, Hibi et al. confirmed that detection of methylated TFPI2 in serum DNA was derived from CRC [7]. Thus, TFPI2 methylation was relevant to tumorigenesis and prognosis of CRC, but there are little strategies were provided to prevent TFPI2 hypermethylation in CRC.
DNA methylation is an epigenetic marker, which is important for controlling gene expression. While ten-eleven translocation (TET) family mediates the sequential oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), then further to 5-formylcytosine and 5-carboxylcytosine, leading to eventual DNA demethylation [8,9,10,11,12]. Among TET family, the most-studied member is TET1. In CRC and some CRC cell lines, mRNA expression of TET1 and global 5hmC level were detected lower than normal tissue or normal colon cells [13,14,15,16,17]; TET1 was capable to react with TSGs by depressing DNA methylation [14, 18], suggesting that enhancement of TET1 expression could be a feasible way of preventing methylation of TSGs of CRC.
ADP-ribosylation is an important post-translational modification of protein. It alters the functional proteins or recruits other proteins by providing a scaffold on the modified proteins and thus regulates several cellular processes. Recently, the function of nuclear ADP-ribosylation in epigenetics became a novel focus [19]. Besides, Ciccarone et al. reported that nuclear poly-ADP-ribosylation was a key positive epigenetic regulator of TET1 transcription by maintaining an active chromatin state of promoter [20]. Interestingly, it was not poly- but mono- or oligo-ADP-ribosylation that was the primary type of histone ADP-ribosylation, while histone poly-ADP-ribosylation was a responsible result to some stress condition [21]; however, the contribution of histone mono-ADP-ribosylation for transcription of TET1 is largely unknown as an epigenetic event.
Since a specific site of histone ADP-ribosylation could cause specific nucleosome structure changes, identification of the target amino acids of histone ADP-ribosylation will further illuminate the interaction between histone and DNA [22]. In previous research, we detected mono-ADP-ribosylation on H3R117 in LoVo cells by liquid chromatography-tandem mass spectrometry (LC-MS/MS), and mutated arginine (R) 117 of H3 to non-ADP-ribosylated alanine (A), as H3R117A LoVo cells, observing depressing proliferation of H3R117A LoVo cells by mutation of H3 mono-ADP-ribosylated R117 [23]. Therefore, we speculated that mono-ADP-ribosylated modification on H3R117 definitely did some contribution to alter chromatin microenvironment of some specific genes and induced subsequently influence malignant biological behavior of cancer cells. However, further research needs to be done.
Thus, in this study, we assessed the effect of mono-ADP-ribosylated H3R117 of LoVo cells on methylation of TSG TFPI2, which was associated with tumorigenesis and prognosis of CRC, and discussed potential epigenetic mechanism involving TET1.
Results
Mono-ADP-ribosylation of H3R117 impacted methylated and hydroxymethylated modification of TSG TFPI2 as well as secretion of TFPI2
DNA of each group was extracted by using Methylated DNA Immunoprecipitation (MeDIP) kit to quantify with size range of DNA (200–1000 base pairs) (Fig. 1). To evaluate effect of mono-ADP-ribosylation of H3R117 on methylated modification level of TFPI2, we used MeDIP-quantitative PCR (qPCR) to measure the enrichment of 5mC on TFPI2 promoter. Inspiringly, quantitation of these observations showed a lower methylation level in H3R117A mutant LoVo cells compared with untreated or empty vector-transfected LoVo cells, accordingly, higher secretion of TFPI2 was detected in H3R117A mutant LoVo cells with respect to other two control groups by enzyme-linked immunosorbent assay (ELISA) (Fig. 1), presenting mono-ADP-ribosylated H3R117 did the contribution to hypermethylated modification of TFPI2 promoter and inhibited secretion of TFPI2 on LoVo cells. We further analyzed hydroxymethylated modification level of TFPI2 promoter by applying hydroxymethylated DNA immunoprecipitation (hMeDIP)-qPCR; interestingly, we found hydroxymethylation of TFPI2 promoter in H3R117A LoVo cells was reduced with respect to control or empty vector-transfected LoVo cells as well (Fig. 1), demonstrating that hydroxymethylation is an independent epigenetic modification of methylation.
Mono-ADP-ribosylation of H3R117 altered methylated and hydroxymethylated modification of TFPI2 promoter DNA as well as secretion of TFPI2. DNA was extracted from cultured cells and sheared into length of 200–1000 bp DNA by sonication. Then, sheared DNA was immunoprecipitated with anti-5-methylcytosine or anti-5-hydroxymethylcytosine. Sample was amplified with primer specific for TFPI2 promoter. For Methylated DNA Immunoprecipitation-quantitative PCR (MeDIP-qPCR), relative fold enrichment was calculated as the ratio of amplification efficiency of the MeDIP sample over that of non-immune IgG as of the following the formula: relative fold enrichment = 2(Ct input − Ct sample)/2(Ct input − Ct IgG). For Hydroxymethylated DNA Immunoprecipitation (hMeDIP)-qPCR, fold enrichment was calculated as the ratio of amplification efficiency of the hMeDIP sample over that of non-immune IgG, fold enrichment % = 2(Ct IgG − Ct sample) × 100%. Since TFPI2 was a secretory protein, enzyme-linked immunosorbent assay (ELISA) kit was used to detect the secretion of TFPI2 in each group, and absorbance was measured by ELISA reader at optical density 450 nm (*P < 0.05 vs control, #P < 0.05 vs empty vector, **P < 0.001 vs control)
Mono-ADP-ribosylation of H3R117 inhibited affinity of TET1 to TFPI2 promoter in LoVo cells by modulating H3K9me3 modification rather than influence of poly-ADP-ribosylation
Sufficient DNA of each group was extracted by using chromatin immunoprecipitation (ChIP) kit to quantify with size range of DNA (200–1000 base pairs) (Fig. 2). Consistent with lower methylation of TFPI2, ChIP assay results revealed that more enrichment of TET1 was observed on TFPI2 promoter in H3R117A LoVo cells compared with control and empty vector-transfected LoVo cells, suggesting that TET1 enriched in TFPI2 promoter and hydroxylated 5mC leading to a decrease of methylation of TFPI2 promoter in H3R117A LoVo cells. Nevertheless, ChIP assay showed no significant difference of poly(ADP-ribose) (PAR) enrichment on the TFPI2 promoter among H3R117A LoVo cells, untreated, and empty vector-transfected LoVo cells (Fig. 3), indicating that mono-ADP-ribosylation of H3R117 did not change poly-ADP-ribosylation of TFPI2 promoter, or, in other words, it precluded the influence of poly-ADP-ribosylation on DNA methylation of TFPI2 promoter.
Chromatin immunoprecipitation (ChIP) for TFPI2 promoter. This figure showed the enrichment of PAR/TET1/H3K4me3/H3K9me3/H3K27me3 on TFPI2 promoter in each group by ChIP assay followed by quantitative PCR analysis. % input = 2% × 2(Ct 2% input sample − Ct IP sample) (**P < 0.001 vs control, ##P < 0.001 vs empty vector, *P < 0.05 vs control, #P < 0.05 vs empty vector)
Considering H3K9me3-marked genes were not targeted for hyper- or hypomethylation by TET proteins [24], we assessed enrichment of H3K9me3 on TFPI2 promoter by ChIP-qPCR. The result showed barely enrichment of H3K9me3 on TFPI2 promoter in H3R117A LoVo cells compared with control and empty vector-transfected LoVo cells (Fig. 3), and demonstrated that loss of H3K9me3 exposed target sites to TET proteins, allowing TET proteins to bind on TFPI2 promoter and hydroxylate 5mC in H3R117A LoVo cells. That was mono-ADP-ribosylation of H3R117 protected DNA hypermethylation from TET1 with modification of H3K9me3 on TFPI2 promoter.
Mono-ADP-ribosylation of H3R117 impacted H3K4me3 and H3K27me3 modifications on TFPI2 promoter in LoVo cells
Zhou et al. reported that loss of TET1 increased histone H3K27me3 causing repression of the target gene E-cadherin in DLD1 colon cancer cells [25]. Accordingly, our data showed that relative lower affinity of TET1 presented higher enrichment of H3K27me3 on TFPI2 promoter in control LoVo cells and empty vector LoVo cells, comparing with H3R117A mutation of LoVo cells (Fig. 3), suggesting that mono-ADP-ribosylation of H3R117 impacted H3K27me3 enrichment on TFPI2 promoter in LoVo cells by inhibiting affinity of TET1.
Putiri et al. proposed that even occupied by TET1, lack of 5hmC appeared commonly at promoters enriched with H3K4me3, due to TET2 played a major role in 5hmC removal on these loci [24]. Thus, to explain reduced hydroxymethylation of TFPI2 promoter on H3R117A mutant LoVo cells occupying with more TET1 enzyme, we detected H3K4me3 modification of TFPI2 promoter. Our data showed a significant increase in the H3K4me3 level of the TFPI2 promoter on H3R117A mutant LoVo cells with respect to mono-ADP-ribosylated H3R117 LoVo cells (Fig. 3), suggesting that, although there is high enrichment of TET1 at the TFPI2 promoter, lower hypohydroxymethylation of TFPI2 promoter was shown in H3R117A mutant LoVo cells, probably owing to the relatively higher enrichment of H3K4me3 on the promoter inducing TET2 to remove 5hmC from the TFPI2 promoter.
Mono-ADP-ribosylation of H3R117 inhibited transcription and expression of TET1
To evaluate the mRNA and protein levels of TET1, we performed Quantitative real-time PCR (RT-qPCR), western blot, and immunofluorescence. Our results showed that, compared with untreated or empty vector-transfected LoVo cells, H3R117A mutant LoVo cells were detected with significantly higher mRNA levels of TET1 (Fig. 4). Accordingly, western blot analysis demonstrated greater TET1 protein level in H3R117A LoVo cells than other two control groups of LoVo cells (Fig. 4), meanwhile, immunofluorescence showed higher average optical density (OD) in the nucleus of H3R117A mutant LoVo cells comparing with control and empty vector group (Fig. 4), suggesting that mono-ADP-ribosylation of H3R117 inhibited expression of TET1 in LoVo cells.
Mono-ADP-ribosylation of H3R117 decreased mRNA and protein expression of TET1. Quantitative PCR was applied for detecting mRNA level of TET1 for each group, while western blot and immunofluorescence were applied for measuring protein expression of TET1 for each group. The grayscale values of the bands of mutant group TET1/H3 were compared with those of the control band TET1/H3 and the ratios were subjected to statistical analysis. For immunofluorescence, average optical density was measured by ImageJ software (*P < 0.05 vs control, **P < 0.001 vs control, ##P < 0.001 vs empty vector)
Mono-ADP-ribosylation of H3R117 influenced DNA methylation but not hydroxymethylation of TET1 promoter
We measured the alters of methylated and hydroxymethylated level of TET1 promoter by MeDIP-qPCR and hMeDIP-qPCR. Consistent with boosted mRNA and protein levels of TET1, 5mC enrichment of TET1 promoter was reduced while 5hmC enrichment of TET1 promoter was not changed in H3R117A LoVo cells with respect to untreated or empty vector-transfected LoVo cells (Fig. 5), illustrating mono-ADP-ribosylation of H3R117 facilitated hypermethylation but not hydroxymethylation of TET1 promoter.
Mono-ADP-ribosylation of H3R117 altered methylated and hydroxymethylated modification of TET1 promoter. Methylated DNA Immunoprecipitation (MeDIP) kit-extracted DNA was immunoprecipitated with anti-5mC or anti-5hmC. Sample was amplified with primer specific for TET1 promoter. For MeDIP-quantitative PCR, relative fold enrichment was calculated as the ratio of amplification efficiency of the MeDIP sample over that of non-immune IgG as of the following the formula: relative fold enrichment = 2(Ct input − Ct sample)/2(Ct input − Ct IgG). For Hydroxymethylated DNA Immunoprecipitation (hMeDIP)-qPCR, fold enrichment was calculated as the ratio of amplification efficiency of the hMeDIP sample over that of non-immune IgG, fold enrichment % = 2(Ct IgG − Ct sample) × 100% (** P < 0.001 vs control, ##P < 0.001 vs empty vector)
Mono-ADP-ribosylation of H3R117 influenced chromatin microenvironment of TET1 promoter
ChIP assay was used to detect the chromatin environment of TET1 promoter. We found that PAR was abundant on TET1 promoter in H3R117A LoVo cells with respected to untreated and empty vector-transfected LoVo cells, suggesting that mono-ADP-ribosylated H3R117 mutation increased poly-ADP-ribosylated modification on TET1 promoter. Meanwhile, increased enrichment of H3K4me3 and decreased enrichment of H3K27me3 were discovered on TET1 promoter in H3R117A LoVo cells, while no differences were detected in enrichment of H3K9me3 among the groups (Fig. 6). These results demonstrated that mono-ADP-ribosylation of H3R117 resulted in chromatin microenvironment of TET1 promoter maintained low local poly-ADP-ribosylation, low H3K4me3 level, and high H3K27me3 level. Interestingly, decreased enrichment of PARP1 on TET1 promoter was discovered in H3R117A LoVo cells (Fig. 6), which seemed contradictory with high enrichment of PAR on TET1 promoter in H3R117A LoVo cells. We would explain the possible reason for this phenomenon in discussion.
Chromatin immunoprecipitation (ChIP) for TET1 promoter. This figure showed the enrichment of PAR/PARP1/H3K4me3/H3K9me3/H3K27me3 on TET1 promoter in each group by ChIP assay followed by quantitative PCR analysis. % input = 2% × 2(Ct 2% input sample − Ct IP sample) (mean %input) ± SD (n = 3) (**P < 0.001 vs control, ##P < 0.001 vs empty vector, *P < 0.05 vs control, #P < 0.05 vs empty vector)
Mono-ADP-ribosylation of H3R117 had no impact on chromatin accessibility of TFPI2 but depressed chromatin accessibility of TET1
Moreover, to explore whether mono-ADP-ribosylation H3R117 impacted chromatin structure at TFPI2 and TET1 promoter, chromatin accessibility assay was applied, and data were presented as the fold enrichment % of amplification efficiency of the Nse-treated DNA sample over that of the No-Nse sample. There was no distinct difference in fold enrichment % at the TFPI2 promoter among H3R117A LoVo cells, untreated, and empty vector-transfected LoVo cells; however, we detected significantly higher fold enrichment % at the TET1 promoter in H3R117A LoVo cells than in control and empty vector-transfected LoVo cells (Fig. 7), indicating that mono-ADP-ribosylation of H3R117 contributed to condensation of TET1 promoter in LoVo cells but not condensation of TFPI2 promoter.
Mono-ADP-ribosylation of H3R117 inhibited chromatin accessibility of TET1 but not TFPI2 promoter. Data were presented as the fold enrichment % of amplification efficiency of the Nse-treated DNA sample over that of the No-Nse sample: fold enrichment % = 2(Ct Nse − Ct No-Nse) × 100% (**P < 0.001 vs control, ##P < 0.001 vs empty vector)
H3R117A mutation reducing methylation of TFPI2 promoter and increasing secretion of TFPI2 via TET1 in LoVo cells
To investigate whether TET1 contributed to attenuation of methylation of TFPI2 promoter in H3R117A mutant LoVo cells, TET1 CRISPR/Cas9 KO plasmid was used to inhibit expression of TET1 in H3R117A mutant LoVo cells. As indicated in Fig. 8, TET1 CRISPR/Cas9 KO plasmid was confirmed efficiently downregulating TET1 mRNA expression. Moreover, raising methylated modification of TFPI2 promoter was presented after transfecting TET1 CRISPR/Cas9 KO plasmid into H3R117A mutant LoVo cells (Fig. 8), which demonstrated that H3R117A mutation cut down methylated modification of TFPI2 promoter depended on increasing of TET1 expression in LoVo cells. Besides, after transfecting TET1 CRISPR/Cas9 KO plasmid into H3R117A mutant LoVo cells the secretion of TFPI2 was detected much lower than H3R117A mutant LoVo cells and control CRISPR/Cas9 group (Fig. 8), suggesting that increasing secretion of TFPI2 in H3R117A mutation LoVo cells due to increasing of TET1 as well.
TET1 knockout (KO) of H3R117A LoVo cells by TET1 CRISPR/Cas9 plasmid boosted methylated modification of TFPI2 promoter and inhibited secretion of TFPI2. Quantitative PCR was applied for confirming TET1 KO after transfection of TET1 CRISPR/Cas9 plasmid, then, DNA was extracted from cultured cells and sheared into length of 200–1000 bp DNA by sonication. Sheared DNA immunoprecipitated with anti-5mC and amplified with primer specific for TFPI2 promoter. Relative fold enrichment was calculated as the ratio of amplification efficiency of the Methylated DNA Immunoprecipitation (MeDIP) sample over that of non-immune IgG as of the following the formula: relative fold enrichment = 2(Ct input − Ct sample)/2(Ct input − Ct IgG). Enzyme-linked immunosorbent assay (ELISA) kit was also used to detected whether secretion of TFPI2 was impacted by TET1 KO in H3R117A LoVo cells, and absorbance was measured by ELISA reader at optical density 450 nm (**P < 0.001 vs control, ##P < 0.001 vs empty vector, *P < 0.05 vs control, #P < 0.05 vs empty vector)
Mono-ADP-ribosylation of H3R117 promoted proliferation of LoVo cells and H3R117A mutation depressed proliferation of LoVo cells depended on TET1
Our previous study confirmedlower proliferation of H3R117A mutant LoVo cells by CCK-8, soft agar cloning, and flow cytometry compared with LoVo cells [23]. In this study, we applied Edu assays to evaluate the role of mono-ADP-ribosylation of H3R117 in increasing proliferation of LoVo cells, further investigated whether TET1 contributed to depression of proliferation in H3R117A mutant LoVo cells. Consistent with our previous research, H3R117A mutant LoVo cells showed lower proliferation comparing with control LoVo cells and empty vector group (Fig. 9). Nevertheless, the proliferation of H3R117A mutant LoVo cells was enhancing again after KO TET1 by transfected with TET1 CRISPR/Cas9 KO plasmid (Fig. 9), indicated that mono-ADP-ribosylation of H3R117 indeed increased proliferation of LoVo cells, and TET1 was facilitate to inhibit proliferation of LoVo cells.
H3R117A mutation depressed proliferation of LoVo cells depending on TET1. Edu assay was applied to detect the consequence of H3R117A mutation on proliferation of LoVo cells, meanwhile, we knockout TET1 in H3R117A LoVo cells to determine the contribution of TET1 to this consequence. The average optical density (AOD) was measured by ImageJ software (*P < 0.05 vs control, #P < 0.05 vs empty vector)
Hypermethylation of TFPI2 was a ubiquitous trend and early event in CRC, and TET1 was positive to TFPI2 expression in normal and malignant tissue
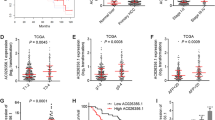
The bioinformatics results showed that hypermethylation of TFPI2 was a common phenomenon in CRC comparing with normal tissue according to the The Cancer Genome Atlas (TCGA), and hypermethylation of TET1 was also detected in colon cancer with respect to normal tissue, but this phenomenon was not observed in rectal cancer (Fig. 10). However, the overall survival was no obvious difference between high TFPI2 or TET1 transcript level and low TFPI2 or TET1 transcript level groups, moreover, different stages of CRC patients appeared no different transcript level of TFPI2 and TET1 (Fig. 10), indicated that hypermethylation of TFPI2 in CRC was an early event. Besides, positive, but not high, correlation of TET1 and TFPI2 expression was detected in normal and malignant tissue according to the TCGA data (Fig. 10), suggesting that TET1 at least partially positively regulated TFPI2 expression.
Bioinformatics analyses for abnormal methylation of TFPI2 and TET1 in colorectal cancer (CRC), as well as correlation and prognosis influence of transcript level of TFPI2 and TET1 of CRC and normal tissue. Methylated modification of TFPI2 and TET1 in normal and CRC was performed using MethHC database, while influence of TFPI2 and TET1 transcript level on overall survival and cancer stage classification of CRC as well as the correlation of these two genes was performed by Gene Expression Profiling Interactive Analysis (the cutoff % of high and low TFPI2/TET1 TPM was 50%)
Discussion
Recently, nuclear poly-ADP-ribosylation had been noticed as an epigenetic marker, due to its influence on DNA methylation, histone modification, and chromatin remodel, all the core histones could be modified by ADP-ribose, but primarily mono-ADP-ribosylation or short oligomers of ADP-ribose [19]. In our previous research, mono-ADP-ribosylation of H3R117 was identified in human colon carcinoma cell line (LoVo cells) by LC-MS/MS, and this post-translational modification might promote the proliferation of colon carcinoma cells in vitro and in vivo, furthermore, H3R117A LoVo cells were successfully constructed to prevent ADP-ribosylation occur on this amino-acid location [23], however, the function of mono-ADP-ribosylated specific amino acid of histone for epigenetics is still unknown.
In this study, we investigated the contribution of mono-ADP-ribosylation of specific amino acid H3R117 to hypermethylation of TSG TFPI2, and discovered mono-ADP-ribosylation of H3R117 facilitated methylation of TSG TFPI2 and inhibited product of TFPI2 protein in LoVo cells. Besides, we found more TET1 enriched on TFPI2 promoter in H3R117A LoVo cells rather than control or empty vector-transfected LoVo cells. Owing to TET1 was the dioxygenases catalyze conversion of 5mC to 5hmC [8]; thus, enrichment of TET1 on TFPI2 promoter generated conversion of 5mC and diminished methylated modification of TFPI2 promoter in H3R117A LoVo cells. On the contrary, mono-ADP-ribosylated H3R117 protected methylation of TFPI2 in LoVo cells by depressing TET1 binding on promoter. Although poly-ADP-ribosylation was associated with DNA methylation [26,27,28], there was no difference of poly-ADP-ribosylation on TFPI2 promoter between mono-ADP-ribosylated H3R117 LoVo cells and H3R117A LoVo cells, suggesting that mono-ADP-ribosylation of H3R117 did not impact local poly-ADP-ribosylation of TFPI2 promoter. Thus, we excluded in influence of poly-ADP-ribosylation on DNA methylation of TFPI2.
On the other hand, we discovered that histone methylation on TFPI2 promoter was also impacted by mono-ADP-ribosylated H3R117. Puriti et al. found that H3K9me3-marked genes were protected from methylation changes resulted from TET1 [24], therefore, relative higher level of H3K9me3 of LoVo cells prevented TET1 binding on TFPI2 promoter DNA, subsequently, maintain hypermethylation of TFPI2 by protecting 5mC from hydroxymethylation, leading to depression of demethylation of TFPI2 promoter in mono-ADP-ribosylated H3R117 LoVo cells. Besides, we observed a strong enrichment of H3K27me3 level on TFPI2 promoter in non-mutated LoVo cells, which was reduced after H3R117A mutation, combining with hypermethylation of TFPI2 promoter in non-mutated LoVo cells and decreased methylation after H3R117A mutation. These findings were in line with previous reports; promoters that were marked with H3K27me3 are more likely to show increased DNA methylation during differentiation and carcinogenesis than those lacking H3K27me3 [29,30,31]; thus, hypermethylation of TFPI2 promoter in mono-ADP-ribosylated H3R117 LoVo cells was also associated with strong enrichment of H2K27me3 at theTFPI2 promoter. However, because of no difference in poly-ADP-ribosylation, the reason for H3R117A mutation leading to change in chromatin microenvironment on TFPI2 promoter is still obscure. Further research needs to be done.
Meanwhile, we examined diminished hydroxymethylated modification of TFPI2 promoter in H3R117A LoVo cells as well. Together with the result of methylation detection on TFPI2 promoter, indicated that 5hmC was an epigenetic marker independent of 5mC, however, the function of 5hmC seemed obscure so far. Recently, considerable evidences had indicated that the presence of 5hmC at the gene promoter negatively regulates gene expression [32,33,34]. Therefore, low hydroxymethylated modification of TFPI2 promoter might another mechanism to reactive TFPI2 in H3R117A LoVo cells. Interestingly, even TFPI2 promoter was enriched for TET1 in H3R117A LoVo cells, a decreased hydroxymethylation at these loci was observed in H3R117A LoVo cells in our study. Although all of TET1, TET2, and TET3 catalyzed conversion of 5mC to 5hmC, however, TET2 or TET3 played a major role in 5hmC removal, moreover, 5hmC removal by TET2 was associated with genes enriched for H3K4me3 [24]. Therefore, our data showed that H3R117A mutation in LoVo cells presented reduced 5hmC level with TET1 occupation on TFPI2 promoter, probably on account of H3K4me3 enrichment inducing 5hmC removal by TET2. Further research needs to be done.
In addition, boosting transcription and expression of TET1 were observed in H3R117A mutant LoVo cells, demonstrating that mono-ADP-ribosylated H3R117 indeed contributed to inhibit TET1 expression in LoVo cells. Besides, abundant enrichment of PAR was measured on TET1 promoter in H3R117A mutant LoVo cells. In other words, our data demonstrated that mono-ADP-ribosylated H3R117 not only changed local poly-ADP-ribosylation of TET1 promoter but also the transcription and expression of TET1. Due to multi-level effects of poly-ADP-ribosylation on epigenetic events [26,27,28], we analyzed chromatin environment on TET1 promoter to explore whether change of local poly-ADP-ribosylation induced by H3R117A mutation impacted relevant epigenetic events on TET1 promoter.
First, DNA methylation was a modulator of gene transcription. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In our research, we found decreasing methylated modification on TET1 promoter in H3R117A mutant LoVo cells versus control LoVo cells. According to Ciccarone’s report that poly-ADP-ribosylation of local chromatin modulated DNA methylation of TET1 gene [20], we speculated that attenuated methylation of TET1 promoter in H3R117A LoVo cells was associated with increased local poly-ADP-ribosylated modification, while mono-ADP-ribosylated H3R117 resulted in abnormal methylation of TET1 gene by reducing poly-ADP-ribosylated modification on TET1 promoter.
Second, the nucleosome limited the accessibility of many regulatory factors. In general, the more accessible the DNA fragments the more likely surrounding genes were actively transcribed. Martinez-Zamudio et al. revealed that histone ADP-ribosylation facilitates gene transcription by destabilizing histone–DNA interaction and increasing the site accessibility of nucleosomal DNA to nucleases [22]. Consistently in our study, enrichment of PAR on TET1 promoter encouraged TET1 transcription by decondensation chromatin of TET1 promoter in H3R117A LoVo cells, while mono-ADP-ribosylated H3R117 played an opposite role to depress transcription of TET1 in non-mutated LoVo cells. On the other hand, chromatin accessibility of TFPI2 promoter was not changed, probably due to no significant difference of PAR on TFPI2 promoter.
In agreement with TET1 transcriptional state, ChIP assay evidenced a raised enrichment of the active transcription marker H3K4me3 on TET1 promoter in H3R117A LoVo cells. Therefore, mono-ADP-ribosylated H3R117 mutation changed original status of low H3K4me3 enrichment on TET1 promoter, and facilitated TET1 transcription in some degrees. Namely, LoVo cells were not enriched for the active transcription mark H3K4me3 on TET1 promoter with mono-ADP-ribosylated H3R117. It was proved that poly-ADP-ribosylation of the histone lysine demethylase KDM5B blocked its binding to chromatin, thus restrained demethylation of H3K4me3 [35]. Hence, we inferred mono-ADP-ribosylated H3R117 decreasing local poly-ADP-ribosylation of TET1 promoter, which allowed activity of KDM5B and diminished H3K4me3 modification on TET1 promoter. Besides, Yamaguchi et al. recently reported that poly-ADP-ribosylation of EZH2 induced PRC2 complex dissociation and EZH2 downregulation, which in turn reduces EZH2-mediated H3K27 trimethylation [36]. This conclusion explained mono-ADP-ribosylated H3R117 mutation resulted in relative low H3K27me3 level with more poly-ADP-ribosylation on TET1 promoter, probably due to increasing poly-ADP-ribosylation impacted function of EZH2, which in turn reduced EZH2-mediated H3K27me3. While H3K27me3 was considered as a repressive transcription mark, thus, low H3K27me3 level of TET1 promoter could be one of the reasons for increasing expression of TET1 in H3R117A LoVo cells.
The evidences above indicated that due to limited poly-ADP-ribosylated modification on TET1 promoter in LoVo cells with mono-ADP-ribosylated H3R117, DNA methylation of TET1 was increased while chromatin accessibility and H3K4me3 level on TET1 promoter were inhibited. These epigenetic events could abate transcription and expression of TET1 in LoVo cells with mono-ADP-ribosylated H3R117.
Interestingly, we found dropped enrichment of PARP1 on TET1 promoter H3R117A LoVo cells, which seemed conflict with increased poly-ADP-ribosylation on this DNA fragments. Nevertheless, PARP1 was not the only PAR polymerase involve in nuclear poly-ADP-ribosylation. PARP2 was also been proved localized in the nucleus and might account for the residual PAR synthesis when PARP1 was deficient [37]. Additionally, Ciccarone et al. showed lower TET1 mRNA level in PARP2-silenced cells rather than PARP1-silenced cells [20]. These evidences suggested that PARP2 played a more important role on transcription of TET1. Thus, we conjectured that it was PARP2 instead of PARP1 mainly responsible for poly-ADP-ribosylation on TET1 promoter in H3R117A LoVo cells. But verification test needs to be done.
To prove the key role of TET1 on modulating methylation of TFPI2 promoter, TET1 was knockout (KO) by TET1 CRISPR/Cas9 KO plasmid in H3R117A LoVo cells. Our results showed that even H3R117A mutation reduced methylation of TFPI2 promoter, after KO TET1 of H3R117A LoVo cells TFPI2 promoter regained methylated modification, suggesting that TET1 was facilitated of demethylation of TFPI2 promoter in LoVo cells. Likewise, secretion of TFPI2 was suppressed in TET1 CRISPR/Cas9 KO plasmid-transfected H3R117A LoVo cells, demonstrating that TET1 enhances secretion of TFPI2. Consistent with our previous study [23] we applied Edu assay also to prove mono-ADP-ribosylated H3R117 promoted proliferation of LoVo cells, besides, we observed proliferation of H3R117A LoVo cells was recovered after KO TET1. Since secretion of TFPI2 was also attenuated after TET1 KO, considering TFPI2 was a tumor suppressor of CRC and had been reported inhibited proliferation [38], we inferred H3R117A inhibited proliferation of LoVo cells by regulating TET1-mediated secretion of TFPI2.
Interestingly, we found that hypermethylation of TFPI2 was prevalent in CRC comparing with normal tissue, but the transcription of TFPI2 seemed no influence on overall survival and staging according to bioinformatics analyses, neither transcription of TET1. These data manifested that hypermethylation of TFPI2 was early event of tumorigenesis, which consistent with Rasmussen’s point that hypermethylated TFPI2 was associated with recurrence and early stage of CRC [6], like a switch of turning on tumor. Thus, reversing hypermethylation of TFPI2 at early stage of cancer, probably an efficient pathway to inhibit tumorigenesis. Bioinformatics analyses was also showed the transcription of these two genes had a positive correlation with each other in CRC and normal tissue, suggesting TET1 at least partially influenced TFPI2 expression in CRC and normal tissue.
In summary, in this study we demonstrated that mono-ADP-ribosylation of H3R117 limited local poly-ADP-ribosylation of TET1 promoter. This limitation impacted following epigenetic events, increased methylation level, and inhibited chromatin accessibility of TET1 promoter as well as H3K4me3 level on TET1 promoter, which reduced transcription and expression of TET1 in LoVo cells. Subsequently, less TET1 were provided to hydroxymethylate 5mC. On the other side, mono-ADP-ribosylation of H3R117 maintained H3K9me3 modification on TFPI2 promoter, which isolated targets of TFPI2 promoter from TET1. Thus, mono-ADP-ribosylation of H3R117 exhausted expression of TET1 and H3K9me3-dependent depressed affinity of TET1 to TFPI2 promoter, impairing demethylation of TET1, coupled with high H3K27me3 level of TFPI2 promoter to recruit DNA methylation, thus contributing to abnormal methylation of TFPI2. Moreover, methylation of TFPI2 was boosted and secretion of TFPI2 was reduced after TET1 KO in H3R117A LoVo cells, further proved effect of TET1 on TFPI2 methylation and expression.
However, since mono-ADP-ribosylation of H3R117 is a genome modification rather than one nucleosome, further research on analysis of epigenetic changes of genome especially TSGs and oncogenes resulting from mono-ADP-ribosylation of H3R117 can shed light on the role of mono-ADP-ribosylation of H3R117 in tumorigenesis. Besides, the impact of mono-ADP-ribosylation of H3R117 on the expression and function of TET2 and TET3 was not investigated in this study. Although we have discussed the possible function of TET2 in hydroxymethylation of TFPI2, further study needs to be done. Nevertheless, our study first highlighted the epigenetic function of mono-ADP-ribosylation of H3R117 on abnormal methylation of TSG TFPI2 by impact TET1, since hypermethyaltion of TFPI2 was an early event in tumorigenesis, selectively target mono-ADP-ribosylation of H3R117 deficiency could be a feasible way to block tumorigenesis of CRC.
Materials and methods
Cells
Human colon adenocarcinoma LoVo cells were kindly obtained from Professor Wei-Xue Tang, Chongqing Medical University (Chongqing, China), while point mutation H3R117 LoVo cells and empty vector-transfected LoVo cells were successfully constructed in our previous study [21]. All these types of LoVo cells were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (Hyclone) at 5% CO2 and 37 °C.
Primary antibodies
The following primary antibodies were used in this study: anti-TET1 antibody (GeneTex, GTX627420), anti-H3 antibody (GeneTex, GTX122148), anti-PAR (Trevigen, 4335-MC-100), anti-PARP1 (Cell Signaling Technology, 9532), anti-H3K9me3 (Cell Signaling Technology, 13969), anti-H3K4me3 (Cell Signaling Technology, 9727), and anti-H3K27me3 (Cell Signaling Technology, 9733).
Primer sequences
Oligonucleotide primers were designed as follows:
TET1 mRNA forward primer TGCCCCACATTGATGAGTATTG,
TET1 mRNA reverse primer AGAGGCGGGTTGGATGATTAC,
Actin mRNA forward primer CACCATTGGCAATGAGCGGTTC,
Actin mRNA reverse primer AGGTCTTTGCGGGTGTCCACGT,
TFPI2 promoter forward primer CTGATTCATGCACGGGGACT,
TFPI2 promoter reverse primer CCCGTCTGGACTACAGGAGA,
TET1 promoter forward primer CAAGTCATGCAGCCCTACCT,
TET1 promoter reverse primer GGCTCCCAGCACAGTCAA.
Western blotting
Western blotting was performed on nuclear protein lysates, which was extracted by Nuclear protein lysates Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China). The concentration of protein was detected by BCA protein assay (Beyotime, China). Then, proteins were separated by weight by SDS-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene fluoride membrane (Millipore, MA, USA). Following pre-incubation with a blocking solution, membranes were incubated in the primary antibody (mouse monoclonal anti-TET1 antibody) at 4 °C (1:500 dilution) overnight. The membranes were then extensively washed, incubated in secondary antibody (1:2000 dilution, ABclonal, China), and were visualized using ChemiDoc XRS (Bio-Rad, USA). Relative protein levels were obtained using Quantity One® software (Bio-Rad, USA) and were normalized to H3 (1:1000 dilution).
Isolation of total RNA and reverse transcription
Total RNA was isolated using CellAmpTM Direct RNA Prep Kit for RT-PCR (Takara, China). Then, complementary DNA was synthesized with the directions of the PrimeScriptTMRT reagent Kit (Perfect Real Time) (Takara, China).
Quantitative PCR
The quantitative real-time PCR followed the directions of the SYBR® Premix Ex Taq™ II (Takara, China), using oligonucleotide primers for human TET1. Actin was used as reference gene for quantification. Quantification was carried out using the geometrical average of 2−△△Ct for TET1 relative to Actin.
MeDIP-qPCR and hMeDIP-qPCR
5mC immunoprecipitation was carried out using the EpiQuik MeDIP Kit (Epigentek, NY, USA) according to the manufacturer’s manual. This kit included a ChIP-grade 5mC antibody and a negative control normal mouse IgG. Briefly, DNA in the cells was extracted and sheared into 200–1000 bp fragments. An aliquot of each sample was set aside as input control, while the remaining portion was subjected to immunoprecipitate with 5mC antibody or normal mouse IgG. DNA was released from the antibody-DNA complex by Proteinase K and purified through the specifically designed Fast-Spin Column. Eluted DNA was amplified with primer specific for TFPI2 promoter or TET1 promoter. Relative fold enrichment presented enrichment of 5mC on TFPI2 promoter: relative fold enrichment = 2(input Ct − sample Ct)/2(input Ct – IgG Ct).
For hMeDIP-qPCR, 5hmC immunoprecipitation was carried out using the EpiQuik hMeDIP Kit (Epigentek), according to the manufacturer’s recommendations. This kit included a positive control DNA fragment, a negative control non-immune IgG, and control primers that can be used with the positive control to demonstrate the enrichment efficacy for hydroxymethylated DNA with the kit reagents and protocol. The positive control DNA containing 5hmC can be immunoprecipitated by a 5hmC antibody but not by a non-immune IgG. Briefly, DNA in the cells was extracted and sheared into 200–1000 bp fragments as described in MeDIP Kit. Sample DNA and positive control DNA were added into the microwell immobilized with 5hmC or IgG antibody, respectively. DNA was released from the antibody-DNA complex by add Proteinase K. Finally, eluted DNA was amplified with primer specific for TFPI2 promoter or TET1 promoter. An input DNA control was generally not needed as the included positive and negative controls could be used for estimating the same objective more accurately. Fold enrichment % was calculated by simply using a ratio of amplification efficiency of the ChIP sample over that of non-immune IgG: fold enrichment % = 2(IgG Ct – Sample Ct) × 100%. The sensitivity of the methods was estimated by analyzing the reference DNA fragment containing 5hmC provided by the kit.
Chromatin accessibility
Chromatin assembly was carried out using the EpiQuik chromatin accessibility assay kit (Epigentek, NY, USA). In this assay, first, cell lysis and chromatin extraction were performed. Then, chromatin was digested with or without a nuclease (Nse) mix, and purified by spin column. Eluted DNA was amplified using qPCR and gene-specific primer for TFPI2 promoter or TET1 promoter. Control primers were provided to determine the successful digestion of the chromatin. The fold enrichment was calculated by the ratio of amplification efficiency of the Nse-treated DNA sample over that of the control sample not treated with nuclease (No-Nse): fold enrichment % = 2(Nse Ct − No-Nse Ct) × 100%. Changes in chromatin structure were identified by the degree of Ct shift between digested and undigested samples.
Chromatin immunoprecipitation
ChIP assays were carried out following the protocol from the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling, 9002). Briefly, cells were treated with 1% formaldehyde for 10 min at room temperature to crosslink proteins to DNA. The chromatin was harvested and digested to a length of approximately 150–900 bp fragments using micrococcal nuclease, then, sonicate lysate with several pulses to break nuclear membrane. An aliquot of each sample was set aside as input control, while the remaining portion was subjected to immunoprecipitation with anti-TET1 (5 μl/IP), anti-PAR (5 μl/IP), anti-PARP1 (2 μl/IP), anti-H3K9me3 (10 μl/IP), anti-H3K4me3 (5 μl/IP), or anti-H3K27me3 (10 μl/IP) overnight at 4 °C, with IgG (5 μl/IP) as negative control. The complex of co-precipitation was captured by ChIP-Grade Protein G Agarose Beads, and chromatin was eluted from antibody/Protein G Beads and reversed crosslinks. Subsequently, DNA was purified and eluted for qPCR. DNA was amplified by PCR using primer pairs designed to amplify TFPI2 promoter or TET1 promoter.
Generation of TET1 CRISPR/Cas9 KO cells
To generate TET1 KO cells, TET1 CRISPR/Cas9 KO Plasmid (h) (Santa Cruz) was transfected into the corresponding H3R117A mutant LoVo cells according to the manufacturer’s instructions, and control CRISPR/Cas9 Plasmid (Santa Cruz)-transfected cells were considered as empty control. Then, 48–72 h after transfection, qPCR analysis was performed to confirm TET1 KO of H3R117A mutant LoVo cells.
Enzyme-linked immunosorbent assay
Quantitative analysis of TFPI2 concentration was respectively conducted by using TFPI2 ELISA kit (Jiangsu Mbbiology Biological Technology Co., Ltd, China) following the manufacturer’s instructions. The absorbance was measured by an ELISA reader (Multiskan FC microplate photometer, Thermo Fisher Scientific) at OD 450 nm.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde at room temperature for 30 min and then permeabilized using 0.5% Triton X-100. Then, cells were washed with cold phosphate-buffered saline to remove free Triton X-100. Subsequently, samples were incubated with primary antibody TET1 at 4 °C overnight. Cells were washed and incubated with Cy3-conjugated secondary antibody (Proteintech, SA00009-1) for 1 h and counterstained with 4′,6-diamidino-2-phenylindole (for nuclear staining). Immunofluorescence was assessed under ZOETM Fluorescent Cell Imager (Bio-Rad) using ImageJ software (version 1.8.0; National Institutes of Health, USA). The assay was repeated in triplicate.
Edu assay
The EdU detection kit (Beyotime, China) was used to evaluate cell proliferation. According to the manufacturer’s protocol, cells were treated with 20 μM EdU for 2 h at 37 °C and fixed with 4% paraformaldehyde at room temperature for 15 min. Following washing with wash buffer three times, cells were treated with 0.5% Triton X-100 for 15 min and stained with Click reaction cocktail for 30 min at room temperature. Following washing with wash buffer, 1× Hoechst 33342 dye was used to incubate cells at room temperature for 10 min. Images were captured using ZOETM Fluorescent Cell Imager (Bio-Rad). Following the merging of the images, the average OD of cells was calculated using ImageJ software (version 1.8.0; National Institutes of Health, USA). The assay was repeated in triplicate.
Bioinformatics analyses
Methylated modification of TFPI2 and TET1 in normal and CRC was performed using MethHC [39], a database of DNA methylation and gene expression in human cancer, which is freely accessible at http://methhc.mbc.nctu.edu.tw/
The influence of TFPI2 and TET1 transcript level on prognosis of CRC patients was performed by Gene Expression Profiling Interactive Analysis (GEPIA), which was developed by Zefang Tang, Chenwei Li, and Boxi Kang of Zhang Lab, Peking University [40]. GEPIA is a newly developed interactive web server for analyzing the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline. GEPIA is available at http://gepia.cancer-pku.cn/.
Statistical analysis
Experiments were replicated at least thrice. Values were presented as mean ± standard deviation (x ± s). Statistical analyses were performed using the GraphPad Prism 6.0 (GraphPad Software, Inc., CA, USA). One-way analysis of variance was adopted, with a significance level defined as P < 0.05.
References
McInnes T, Zou D, Rao DS, Munro FM, Phillips VL, McCall JL. et al. Genome-wide methylation analysis identifies a core set of hypermethylated genes in CIMP-H colorectal cancer. BMC Cancer. 2017;17:228.
Li Y-F, Hsiao Y-H, Lai Y-H, Chen Y-C, Chen Y-J, Chou J-L. et al. DNA methylation profiles and biomarkers of oral squamous cell carcinoma. Epigenetics. 2015;10:229–36.
Liu Z, Zhang J, Gao Y, Pei L, Zhou J, Gu L. et al. Large-scale characterization of DNA methylation changes in human gastric carcinomas with and without metastasis. Clin Cancer Res. 2014;20:4598–612.
Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, et al. Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett. 2011;311:96–100.
Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–9.
Rasmussen SL, Krarup HB, Sunesen KG, Pedersen IS, Madsen PH, Thorlacius-Ussing O. Hypermethylated DNA as a biomarker for colorectal cancer: a systematic review. Colorectal Dis. 2016;18:549–61.
Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, et al. Methylation of TFPI2 no longer detected in the serum DNA of colorectal cancer patients after curative surgery. Anticancer Res. 2012;32:787–90.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3.
Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–8.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7.
Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, et al. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013;14:R91.
Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670–6.
Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: an epigenetic mark frequently deregulated in cancer. Biochim Biophys Acta. 2015;1855:144–54.
Rawłuszko-Wieczorek AA, Siera A, Horbacka K, Horst N, Krokowicz P, Jagodziński PP. Clinical significance of DNA methylation mRNA levels of TET family members in colorectal cancer. J Cancer Res Clin Oncol. 2015;141:1379–92.
Ichimura N, Shinjo K, An B, Shimizu Y, Yamao K, Ohka F, et al. Aberrant TET1 methylation closely associated with CpG island methylator phenotype in colorectal cancer. Cancer Prev Res (Phila). 2015;8:702–11.
Neri F, Dettori D, Incarnato D, Krepelova A, Rapelli S, Maldotti M, et al. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene. 2015;34:4168–76.
Li Lili, Li Chen, Mao Haitao, Du Zhenfang, Chan WaiYee, Murray Paul, et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci Rep. 2016;6:26591.
Hottiger MO. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem. 2015;84:227–63.
Ciccarone F, Valentini E, Bacalini MG, Zampieri M, Calabrese R, Guastafierro T, et al. Poly(ADP-ribosyl)ation is involved in the epigenetic control of TET1 gene transcription. Oncotarget. 2014;5:10356–67.
Burzio L, Riquelme P, Koide S. ADP ribosylation of rat liver nucleosomal core histones. J Biol Chem. 1979;254:3029–37.
Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol. 2012;32:2490–502.
Ling F, Tang Y, Li M, Li QS, Li X, Yang L, et al. Mono-ADP-ribosylation of histone 3 at arginine-117 promotes proliferation through its interaction with P300. Oncotarget. 2017;8:72773–87.
Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi JH, et al. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15:R81.
Zhou Z, Zhang HS, Liu Y, Zhang ZG, Du GY, Li H, et al. Loss of TET1 facilitates DLD1 colon cancer cell migration via H3K27me3-mediated down-regulation of E-cadherin. J Cell Physiol. 2018;233:1359–69.
Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, et al. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS ONE. 2009;4:e4717.
Zampieri M, Guastafierro T, Calabrese R, Ciccarone F, Bacalini MG, Reale A, et al. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J. 2012;441:645–52.
Nocchi L, Tomasetti M, Amati M, Neuzil J, Santarelli L, Saccucci F. Thrombomodulin is silenced in malignant mesothelioma by a poly(ADP-ribose) polymerase-1-mediated epigenetic mechanism. J Biol Chem. 2011;286:19478–88.
Thomson JP, Ottaviano R, Unterberger E, Lempiäinen H, Muller A, Terranova R, et al. Loss of Tet1 associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016;76:3097–108.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:23223–6.
Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42.
Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genomewide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–64.
Robertson J, Robertson AB, Klungland A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochem Biophys Res Commun. 2011;411:40–3.
Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genomewide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84.
Krishnakumar R, Kraus W. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–49.
Yamaguchi H, Du Y, Nakai K, Ding M, Chang SS, Hsu JL, et al. EZH2 contributes to the response to PARP inhibitors through its PARP-mediated poly-ADP ribosylation in breast cancer. Oncogene. 2018;37:208–17.
Am´e JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, et al. PARP-2, a novel mammalian DNA damage–dependent poly(ADP-ribose) polymerase. Biol Chem. 1999;274:17860–8.
Gao S, Lin Z, Li C, Wang Y, Yang L, Zou B, et al. TFPI2AS1, a novel lncRNA that inhibits cell proliferation and migration in lung cancer. Cell Cycle. 2017;16:2249–58.
Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL, et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856–61.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102.
Acknowledgements
We gratefully acknowledge Professor Wei-Xue Tang (Chongqing, China) for his gift of LoVo cell line. This work was supported by the Science and Technology Plan Projects of Yuzhong District in Chongqing [grant numbers 20170409 and 20140106], and the Scientific Research Foundation of Chongqing Medical University [grant number 201413].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Tang, Y., Li, Q. et al. Mono-ADP-ribosylation of H3R117 traps 5mC hydroxylase TET1 to impair demethylation of tumor suppressor gene TFPI2. Oncogene 38, 3488–3503 (2019). https://doi.org/10.1038/s41388-018-0671-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0671-8