Abstract
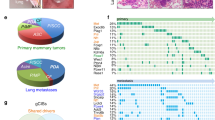
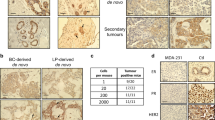
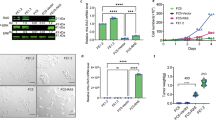
Copy number gains, point mutations and epigenetic silencing events are increasingly observed in genes encoding elements of the Ras/Raf/MEK/ERK signaling axis in human breast cancer. The three Raf kinases A-Raf, B-Raf, and Raf-1 have an important role as gatekeepers in ERK pathway activation and are often dysregulated by somatic alterations of their genes or by the aberrant activity of receptor tyrosine kinases (RTKs) and Ras-GTPases. B-Raf represents the most potent Raf isoform and a critical effector downstream of RTKs and RAS proteins. Aberrant RTK signaling is mimicked by the polyoma middle T antigen (PyMT), which activates various oncogenic signaling pathways, incl. the RAS/ERK axis, in a similar manner as RTKs in human breast cancer. Mammary epithelial cell directed expression of PyMT in mice by the MMTV-PyMT transgene induces mammary hyperplasia progressing over adenoma to metastatic breast cancer with an almost complete penetrance. To understand the functional role of B-Raf in this model for luminal type B breast cancer, we crossed MMTV-PyMT mice with animals that either lack B-Raf expression in the mammary gland or express the signaling impaired B-RafAVKA mutant. The AVKA mutation prevents phosphorylation of T599 and S602 in the B-Raf activation loop and thereby activation of the kinase by upstream signals. We demonstrate for the first time that B-Raf expression and activation is important for tumor initiation in vivo as well as for lung metastasis. Isogenic tumor cell lines generated from conditional Braf knock-out or knock-in mice displayed a reduction in EGF-induced ERK pathway activity as well as in proliferation and invasive growth in three-dimensional matrigel cultures. Our results suggest that B-Raf, which has been hardly studied in the context of breast cancer, represents a critical effector of the PyMT oncoprotein and invite for an assessment of its functional role in human breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Ha JR, Siegel PM, Ursini-Siegel J. The tyrosine kinome dictates breast cancer heterogeneity and therapeutic responsiveness. J Cell Biochem. 2016;117:1971–90.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4.
McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, et al. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–78.
Sears R, Gray JW. Epigenomic inactivation of RasGAPs activates RAS signaling in a subset of luminal B breast cancers. Cancer Discov. 2017;7:131–3.
Olsen SN, Wronski A, Castano Z, Dake B, Malone C, De Raedt T, et al. Loss of RasGAP tumor suppressors underlies the aggressive nature of luminal B breast cancers. Cancer Discov. 2017;7:202–17.
Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13:928–42.
Little AS, Smith PD, Cook SJ. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene. 2013;32:1207–15.
Röring M, Brummer T. Aberrant B-raf signaling in human cancer—10 years from bench to bedside. Crit Rev Oncog. 2012;17:97–121.
Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21.
Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5.
Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30.
Röring M, Herr R, Fiala GJ, Heilmann K, Braun S, Eisenhardt AE, et al. Distinct requirement for an intact dimer interface in wild-type, V600E and kinase-dead B-Raf signalling. EMBO J. 2012;31:2629–47.
Yaktapour N, Meiss F, Mastroianni J, Zenz T, Andrlova H, Mathew NR, et al. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest. 2014;124:5074–84.
Desideri E, Cavallo AL, Baccarini M. Alike but different: RAF paralogs and their signaling outputs. Cell. 2015;161:967–70.
Wojnowski L, Stancato LF, Larner AC, Rapp UR, Zimmer A. Overlapping and specific functions of Braf and Craf-1 proto-oncogenes during mouse embryogenesis. Mech Dev. 2000;91:97–104.
Galabova-Kovacs G, Catalanotti F, Matzen D, Reyes GX, Zezula J, Herbst R, et al. Essential role of B-Raf in oligodendrocyte maturation and myelination during postnatal central nervous system development. J Cell Biol. 2008;180:947–55.
Galabova-Kovacs G, Matzen D, Piazzolla D, Meissl K, Plyushch T, Chen AP, et al. Essential role of B-Raf in ERK activation during extraembryonic development. Proc Natl Acad Sci USA. 2006;103:1325–30.
Zhong J, Li X, McNamee C, Chen AP, Baccarini M, Snider WD. Raf kinase signaling functions in sensory neuron differentiation and axon growth in vivo. Nat Neurosci. 2007;10:598–607.
Pritchard CA, Samuels ML, Bosch E, McMahon M. Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol Cell Biol. 1995;15:6430–42.
Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–67.
Köhler M, Brummer T. B-Raf activation loop phosphorylation revisited. Cell Cycle. 2016;15:1171–3.
Köhler M, Röring M, Schorch B, Heilmann K, Stickel N, Fiala GJ, et al. Activation loop phosphorylation regulates B-Raf in vivo and transformation by B-Raf mutants. EMBO J. 2016;35:143–61.
Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–45.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Ali NA, Wu J, Hochgrafe F, Chan H, Nair R, Ye S, et al. Profiling the tyrosine phosphoproteome of different mouse mammary tumour models reveals distinct, model-specific signalling networks and conserved oncogenic pathways. Breast Cancer Res. 2014;16:437.
Bengsch F, Buck A, Gunther SC, Seiz JR, Tacke M, Pfeifer D, et al. Cell type-dependent pathogenic functions of overexpressed human cathepsin B in murine breast cancer progression. Oncogene. 2014;33:4474–84.
Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73:542–63.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76.
Dilworth SM. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat Rev Cancer. 2002;2:951–6.
Smith BA, Shelton DN, Kieffer C, Milash B, Usary J, Perou CM, et al. Targeting the PyMT oncogene to diverse mammary cell populations enhances tumor heterogeneity and generates rare breast cancer subtypes. Genes Cancer. 2012;3:550–63.
Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26.
Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212.
Ben-David U, Ha G, Khadka P, Jin X, Wong B, Franke L, et al. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat Commun. 2016;7:12160.
Chen AP, Ohno M, Giese KP, Kuhn R, Chen RL, Silva AJ. Forebrain-specific knockout of B-raf kinase leads to deficits in hippocampal long-term potentiation, learning, and memory. J Neurosci Res. 2006;83:28–38.
Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–30.
Marcotte R, Smith HW, Sanguin-Gendreau V, McDonough RV, Muller WJ. Mammary epithelial-specific disruption of c-Src impairs cell cycle progression and tumorigenesis. Proc Natl Acad Sci USA. 2012;109:2808–13.
Lan L, Holland JD, Qi J, Grosskopf S, Rademann J, Vogel R, et al. Shp2 signaling suppresses senescence in PyMT-induced mammary gland cancer in mice. EMBO J. 2015;34:1493–508.
Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–86.
Haug S, Schnerch D, Halbach S, Mastroianni J, Dumit VI, Follo M, et al. Metadherin exon 11 skipping variant enhances metastatic spread of ovarian cancer. Int J Cancer. 2015;136:2328–40.
Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–64.
Gillies TE, Pargett M, Minguet M, Davies AE, Albeck JG. Linear integration of ERK activity predominates over persistence detection in Fra-1 regulation. Cell Syst. 2017;5:549–63 e545.
Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. Embo J. 2000;19:5429–39.
Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJ, et al. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013;154:1036–46.
Thevakumaran N, Lavoie H, Critton DA, Tebben A, Marinier A, Sicheri F, et al. Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation. Nat Struct Mol Biol. 2015;22:37–43.
O’Toole SA, Beith JM, Millar EK, West R, McLean A, Cazet A, et al. Therapeutic targets in triple negative breast cancer. J Clin Pathol. 2013;66:530–42.
Adeyinka A, Nui Y, Cherlet T, Snell L, Watson PH, Murphy LC. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin Cancer Res. 2002;8:1747–53.
Whyte J, Bergin O, Bianchi A, McNally S, Martin F. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 2009;11:209.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67.
Santarpia L, Qi Y, Stemke-Hale K, Wang B, Young EJ, Booser DJ, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012;134:333–43.
Tilch E, Seidens T, Cocciardi S, Reid LE, Byrne D, Simpson PT, et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat. 2014;143:385–92.
Aceto N, Sausgruber N, Brinkhaus H, Gaidatzis D, Martiny-Baron G, Mazzarol G, et al. Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat Med. 2012;18:529–37.
Bentires-Alj M, Gil SG, Chan R, Wang ZC, Wang Y, Imanaka N, et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–21.
Jacob LS, Vanharanta S, Obenauf AC, Pirun M, Viale A, Socci ND, et al. Metastatic competence can emerge with selection of preexisting oncogenic alleles without a need of new mutations. Cancer Res. 2015;75:3713–9.
Herr R, Wohrle FU, Danke C, Berens C, Brummer T. A novel MCF-10A line allowing conditional oncogene expression in 3D culture. Cell Commun Signal. 2011;9:17.
Pearson GW, Hunter T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J Cell Biol. 2007;179:1555–67.
Beliveau A, Mott JD, Lo A, Chen EI, Koller AA, Yaswen P, et al. Raf-induced MMP9 disrupts tissue architecture of human breast cells in three-dimensional culture and is necessary for tumor growth in vivo. Genes Dev. 2010;24:2800–11.
Liu H, Murphy CJ, Karreth FA, Emdal KB, White FM, Elemento O, et al. Identifying and targeting sporadic oncogenic genetic aberrations in mouse models of triple-negative breast cancer. Cancer Discov. 2018;8:354–69.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91.
Varga A, Ehrenreiter K, Aschenbrenner B, Kocieniewski P, Kochanczyk M, Lipniacki T, et al. RAF1/BRAF dimerization integrates the signal from RAS to ERK and ROKalpha. Sci Signal. 2017;10:eaai8482.
Kamata T, Hussain J, Giblett S, Hayward R, Marais R, Pritchard C. BRAF inactivation drives aneuploidy by deregulating CRAF. Cancer Res. 2010;70:8475–86.
Nieto P, Ambrogio C, Esteban-Burgos L, Gomez-Lopez G, Blasco MT, Yao Z, et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature. 2017;548:239–43.
Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, et al. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95.
Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, Davies L, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015;27:85–96.
Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526:583–6.
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13:828–51.
Hillebrand LE, Bengsch F, Hochrein J, Hulsdunker J, Bender J, Follo M, et al. Proteolysis—a characteristic of tumor-initiating cells in murine metastatic breast cancer. Oncotarget. 2016;7:58244–60.
Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Muller S, et al. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci USA. 2010;107:2497–502.
Sevenich L, Werner F, Gajda M, Schurigt U, Sieber C, Muller S, et al. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene. 2011;30:54–64.
Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc. 2012;7:374–93.
Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29:79–83.
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) via the Collaborative Research Centre 850 (projects B4; B7, and C6), the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS), the German Cancer Consortium (DKTK, L627) and the Spemann Graduate School for Biology and Medicine (GSC-4; SGBM) via fellowships to MK, SH, NK, CK. and NR. TB was also supported by the Emmy-Noether and Heisenberg programs of the DFG. We would also like to thank Chris Ormandy for helpful discussions, Ricarda Herr for advice, and Nicole Klemm and Ulrike Reif for expert technical assistance.
Author contributions
MK, TR, and TB designed and conceived experiments, conducted data interpretation and wrote the manuscript. MK, SE, SH, ML, UB, NR, SC, CS, FMU, NK, KR, and SB performed experimental work and data interpretation. CP and RZ contributed to experimental design and data interpretation. TR and TB supervised research and project planning.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Köhler, M., Ehrenfeld, S., Halbach, S. et al. B-Raf deficiency impairs tumor initiation and progression in a murine breast cancer model. Oncogene 38, 1324–1339 (2019). https://doi.org/10.1038/s41388-018-0663-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0663-8
This article is cited by
-
Cathepsin D deficiency in mammary epithelium transiently stalls breast cancer by interference with mTORC1 signaling
Nature Communications (2020)