Abstract
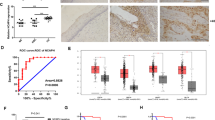
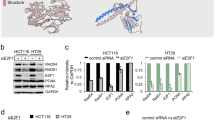
High-risk human papillomaviruses (HPVs) constitutively activate ataxia telangiectasia mutated (ATM) and ataxia telangiectasia- and Rad3-related (ATR) DNA damage repair pathways for viral genome amplification. HPVs activate these pathways through the immune regulator STAT-5. For the ATR pathway, STAT-5 increases expression of the topoisomerase IIβ-binding protein 1 (TopBP1), a scaffold protein that binds ATR and recruits it to sites of DNA damage. TopBP1 also acts as a transcriptional regulator, and we investigated how this activity influenced the HPV life cycle. We determined that TopBP1 levels are increased in cervical intraepithelial neoplasias as well as cervical carcinomas, consistent with studies in HPV-positive cell lines. Suppression of TopBP1 by shRNAs impairs HPV genome amplification and activation of the ATR pathway but does not affect the total levels of ATR and CHK1. In contrast, knockdown reduces the expression of other DNA damage factors such as RAD51 and Mre11 but not BRCA2 or NBS1. Interestingly, TopBP1 positively regulates the expression of E2F1, a TopBP1-binding partner, and p73 in HPV-positive cells in contrast to its effects in other cell types. TopBP1 transcriptional activity is regulated by AKT, and treatment with AKT inhibitors suppresses expression of E2F1 and p73 without interfering with ATR signaling. Importantly, the levels of p73 are elevated in HPV-positive cells and its knockdown impairs HPV genome amplification. This demonstrates that p73, like p63 and p53, is an important regulator of the HPV life cycle that is controlled by the transcriptional activating properties of the multifunctional TopBP1 protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hausen zur H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50.
Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97.
Spurgeon ME, Lambert PF. Human papillomavirus and the stroma: bidirectional crosstalk during the virus life cycle and carcinogenesis. Viruses. 2017;9:219.
Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70.
Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60.
Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–28.
McBride AA. The papillomavirus E2 proteins. Virology. 2013;445:57–79.
DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114.
Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80–98.
Munger K, Howley PM. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–28.
Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci: a J Virtual Libr. 2008;13:1003–17.
Wallace NA, Galloway DA. Novel functions of the human papillomavirus e6 oncoproteins. Annu Rev Virol. 2015;2:403–23.
Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–54.
Thomas M, Narayan N, Pim D, Tomaic V, Massimi P, Nagasaka K, et al. Human papillomaviruses, cervical cancer and cell polarity. Oncogene. 2008;27:7018–30.
Conway MJ, Meyers C. Replication and assembly of human papillomaviruses. J Dent Res. 2009;88:307–17.
Hong S-Y. DNA damage response is hijacked by human papillomaviruses to complete their life cycle. J Zhejiang Univ Sci B. 2017;18:215–32.
Ai W, Narahari J, Roman A. Yin yang 1 negatively regulates the differentiation-specific E1 promoter of human papillomavirus type 6. J Virol. 2000;74:5198–205.
Hartley KA, Alexander KA. Human TATA binding protein inhibits human papillomavirus type 11 DNA replication by antagonizing E1-E2 protein complex formation on the viral origin of replication. J Virol. 2002;76:5014–23.
Offord EA, Beard P. A member of the activator protein 1 family found in keratinocytes but not in fibroblasts required for transcription from a human papillomavirus type 18 promoter. J Virol. 1990;64:4792–8.
O’Connor M, Bernard HU. Oct-1 activates the epithelial-specific enhancer of human papillomavirus type 16 via a synergistic interaction with NFI at a conserved composite regulatory element. Virology. 1995;207:77–88.
Stunkel W, Bernard HU. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol. 1999;73:1918–30.
Gunasekharan V, Hache G, Laimins L. Differentiation-dependent changes in levels of C/EBPbeta repressors and activators regulate human papillomavirus type 31 late gene expression. J Virol. 2012;86:5393–8.
Gunasekharan VK, Li Y, Andrade J, Laimins LA. Post-transcriptional regulation of KLF4 by high-risk human papillomaviruses is necessary for the differentiation-dependent viral life cycle. PLoS Pathog. 2016;12:e1005747.
Zhang W, Hong S, Maniar KP, Cheng S, Jie C, Rademaker AW et al. KLF13 regulates the differentiation-dependent human papillomavirus life cycle in keratinocytes through STAT5 and IL-8. Oncogene 2016. https://doi.org/10.1038/onc.2016.97.
Hong S, Laimins LA. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog. 2013;9:e1003295.
Hong S, Cheng S, Iovane A, Laimins LA. STAT-5 regulates transcription of the topoisomerase iiβ-binding protein 1 (topbp1) gene to activate the ATR pathway and promote human papillomavirus replication. mBio. 2015;6:e02006–15.
Edwards TG, Helmus MJ, Koeller K, Bashkin JK, Fisher C. Human papillomavirus episome stability is reduced by aphidicolin and controlled by DNA damage response pathways. J Virol. 2013;87:3979–89.
Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605.
Kidiyoor GR, Kumar A, Foiani M. ATR-mediated regulation of nuclear and cellular plasticity. DNA Repair (Amst) 2016. https://doi.org/10.1016/j.dnarep.2016.05.020.
Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–304.
Liu K, Paik JC, Wang B, Lin F-T, Lin W-C. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–807.
Liu K, Ling S, Lin W-C. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–81.
Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 2008;27:2851–61.
Liu K, Graves JD, Scott JD, Li R, Lin W-C. Akt switches TopBP1 function from checkpoint activation to transcriptional regulation through phosphoserine binding-mediated oligomerization. Mol Cell Biol. 2013;33:4685–4700.
Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55.
Mehta K, Laimins L. Humanpapillomaviruses preferentially recruit DNA repair factors to viral genomes for rapid repair and amplification. mBio. 2018;9:e00064–18.
Liu K, Bellam N, Lin H-Y, Wang B, Stockard CR, Grizzle WE, et al. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol Cell Biol. 2009;29:2673–93.
Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 2002;16:245–56.
Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–99.
Chappell WH, Gautam D, Ok ST, Johnson BA, Anacker DC, Moody CA. Homologous recombination repair factors Rad51 and BRCA1 are necessary for productive replication of human papillomavirus 31. J Virol. 2015;90:2639–52.
Mighty KK, Laimins LA. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J Virol. 2011;85:8863–9.
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505.
Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–52.
Lorenz LD, Rivera Cardona J, Lambert PF. Inactivation of p53 rescues the maintenance of high risk HPV DNA genomes deficient in expression of E6. PLoS Pathog. 2013;9:e1003717.
Reiser J, Hurst J, Voges M, Krauss P, Münch P, Iftner T, et al. High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J Virol. 2011;85:11372–80.
Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–8.
Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–71.
Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4.
Donaldson MM, Mackintosh LJ, Bodily JM, Dornan ES, Laimins LA, Morgan IM An interaction between human papillomavirus 16 E2 and TopBP1is required for optimum viral DNA replication and episomal genome establishment. J Virol 2012. https://doi.org/10.1128/JVI.01002-12.
Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–21.
Stiewe T, Pützer BM. Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ. 2002;9:237–45.
Fontemaggi G, Kela I, Amariglio N, Rechavi G, Krishnamurthy J, Strano S, et al. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J Biol Chem. 2002;277:43359–68.
Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–60.
Niklison-Chirou MV, Killick R, Knight RA, Nicotera P, Melino G, Agostini M. How does p73 cause neuronal defects? Mol Neurobiol. 2016;53:4509–20.
Wang Z, Sturgis EM, Guo W, Song X, Zhang F, Xu L, et al. Association of combined p73 and p53 genetic variants with tumor HPV16-positive oropharyngeal cancer. PLoS ONE. 2012;7:e35522.
Accardi R, Dong W, Smet A, Cui R, Hautefeuille A, Gabet A-S, et al. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 2006;7:334–40.
Pacini L, Savini C, Ghittoni R, Saidj D, Lamartine J, Hasan UA, et al. Downregulation of toll-like receptor 9 expression by beta human papillomavirus 38 and implications for cell cycle control. J Virol. 2015;89:11396–405.
Habiger C, Jäger G, Walter M, Iftner T, Stubenrauch F. Interferon kappa inhibits human papillomavirus 31 transcription by inducing Sp100 proteins. J Virol. 2016;90:694–704.
Fehrmann F, Laimins LA. Human papillomavirus type 31 life cycle: methods for study using tissue culture models. Methods Mol Biol. 2005;292:317–30.
Hong S, Mehta KP, Laimins LA. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J Virol. 2011;85:9486–94.
Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15:1268–79.
Andrews S FastQC: a quality control tool for high throughput sequence data. 2010.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97.
Yu G, Wang L-G, Han Y, He Q-Y clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. https://homeliebertpubcom/omi 2012; 16: 284–7.
Acknowledgements
We thank the members of Laimins Lab for their help and comments.
Funding
This work is supported by grants from NCI (CA059655 and CA142861) to L.A.L., ACS grant IRG-15-173-21 to S.H., and National Natural Scientific Foundation of China (No. 81702552) to J.X.
Author contributions
L.A.L.: concepts, experimental design, data interpretation, and writing of manuscript. S.H.: concepts, perform experiments, experimental design, data acquisition, analysis, and writing of manuscript. J.A., Y.L.: data analysis. J.X., P.H., P.K.: perform experiments and data acquisition.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Hong, S., Xu, J., Li, Y. et al. Topoisomerase IIβ-binding protein 1 activates expression of E2F1 and p73 in HPV-positive cells for genome amplification upon epithelial differentiation. Oncogene 38, 3274–3287 (2019). https://doi.org/10.1038/s41388-018-0633-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0633-1