Abstract
Yes-associated protein 1 (YAP1) exerts significant effects in various malignancies. However, the oncogenic role of YAP1 remains controversial, and the mechanism by which YAP1 regulates non-coding RNAs is still largely unknown. The present study aimed to assess the effect of YAP1 on the malignant behaviors of colorectal carcinoma (CRC) and explore the underlying regulatory mechanism of the YAP1–MALAT1–miR-126-5p axis. YAP1 was highly expressed in CRC tissues as assessed by GSE20916 and its expression was negatively correlated with overall survival in 83 CRC cases. Meanwhile, YAP1 promoted proliferation, invasion, and migration in colon cancer cells, in vitro and in vivo. MALAT1 was obviously expressed, with differential expression of 11 lncRNAs in HCT116 cells after transfection with siYAP1 or si-Ctl. Based on bioinformatics prediction, immunoprecipitation (IP), and chromatin immunoprecipitation (ChIP), the interaction of YAP1 with TCF4/β-catenin was regulated by MALAT1. Bioinformatics prediction, dual luciferase assay, RNA-IP, and RNA pull-down assay demonstrated that YAP1-induced MALAT1 promoted the expression of metastasis-associated molecules such as VEGFA, SLUG, and TWIST, by sponging miR-126-5p in CRC. These findings indicated that the YAP1–MALAT1–miR-126-5p axis could control angiogenesis and epithelial–mesenchymal transition in CRC, providing potential biomarkers and therapeutic targets for CRC.
Similar content being viewed by others
Introduction
Colorectal carcinoma (CRC) remains one of the most common malignancies worldwide, and is the sixth primary cause of death induced by cancer in China [1, 2]. Moreover, the occurrence of CRC metastasis predicted a bad prognosis [3]. Thus, identifying metastasizing biomarkers in the early CRC stage would facilitate CRC diagnosis as well as the development of therapeutic targets.
Long non-coding RNAs (lncRNAs) are transcribed RNA molecules >200 nucleotides in length [4]. With the development of high sequencing technologies, an increasing number of lncRNAs have been shown to be involved in various biological functions, including tumor proliferation, apoptosis, invasion, metastasis, stemness, metabolism, and differentiation [5,6,7]. In CRC, recent studies reported that lncRNAs were involved in regulating tumor progression [8,9,10]. Moreover, increasing lncRNAs showed a potential to be diagnostic or prognostic markers [11,12,13,14]. However, the underlying mechanism of lncRNAs involved in CRC metastasis and clinical significance of lncRNAs in CRC remain largely unknown.
As we knew, YAP1 showed onco-roles in many cancer [15, 16]. Our previous findings reported that miR-590-5p-associated YAP1 downregulation results in declined CRC tumorigenesis, which illustrated that YAP1 as an oncogene was involved in CRC progression [17]. YAP1 was reported to regulate the maintain stemness, EMT process of cancer [18,19,20]. Moreover, there are a few studies on the regulation relationship between YAP1 and lncRNAs. In pancreatic cancer, Zhou et al. have found that lncRNA MALAT1 is upregulated and the results of si-MALAT1 showed a decline of YAP1 expression [21]. Meanwhile, the fact of lncRNA regulating YAP1 was proved by sponging miR-18a in breast cancer cells. Similarly, lncRNA LUST could directly interact with β-catenin for facilitating it to occupy more promoters related with WNT-signaling target genes including YAP1 in colon cancer stem-like cells [22]. Interestingly, contrary results displayed that si-YAP1 experiment results in a decreasing lncRNA H19 and si-H19 had no effect on YAP1 expression, but no further study [23]. These studies implied that there is a regulation relationship between YAP1 and lncRNAs in cancer. In this study, the role and mechanism of the YAP1 regulating lncRNA were explored in CRC. Based on a series of novel findings, this putative therapeutic target has a potential role in CRC diagnosis and treatment.
Results
YAP1 is an upregulated oncogene in CRC
To assess the role of YAP1 in CRC, we first quantitated YAP1 gene expression levels in CRC biopsies and normal colorectal tissues using a microarray dataset, GSE20916, containing 24 normal colorectal tissues and 81 CRC tissue biopsies, with the Affymetrix HG U133 Plus 2.0 GeneChip platform. Significant analysis of microarray (SAM) [24] was carried out between normal colorectal and CRC tissue samples, and YAP1 was found to be upregulated in CRC samples compared with normal colorectal specimens (p < 0.001) (Fig. 1a). YAP1 overexpression in CRC tissues was confirmed by qRT-PCR and immunohistochemistry (Fig. 1b, c). In addition, the patients were divided into low and high expression groups based on YAP1 mean values. Univariate analysis showed that YAP1 expression was associated with TNM stage, lymphatic metastasis, and distant metastasis (all p < 0.05; Table S1). However, YAP1 expression was not associated with age, tumor location, tumor size, tumor general type, and tumor grade (all p > 0.05). Interestingly, patients with low YAP1 levels survived longer than those with high YAP1 expression (log-rank test, p < 0.05; Fig. 1d). These findings suggested that low YAP1 expression had a positive effect in the prognostic prediction of overall survival (OS) in CRC patients (Table S1). YAP1 was upregulated in colon cancer cells compared with normal colon mucosal cells (NCM460; figure S1a). The results for YAP1 depletion efficiency in vivo and in vitro are shown in figure S4. Subsequently, clonogenic and cell proliferation assays using CCK-8 and EdU staining showed that YAP1 knockdown inhibited HCT116 cell proliferation compared with the negative control group (all p < 0.05; Fig. 1e–g). In addition, Transwell and wound-healing assays showed that YAP1 knockdown suppressed the ability of invasion and migration of HCT116 cells, compared with the negative control group (all p < 0.05; Fig. 1h, i) and vice versa. The above results suggested that YAP1 acted as an oncogene in colon cancer cells in vitro. To assess the biological function of YAP1 in vivo, a nude mice xenograft model was established by subcutaneously injecting HCT116 cells. After 7 days, the subcutaneous tumors were discontinuously injected with RNAi-YAP1 (siYAP1 agomir) or agomir control for 28 days. Tumor volumes and weights in the siYAP1 agomir group were reduced compared with those of the agomir control group (Fig. 1j). YAP1 expression levels in subcutaneous tumors were lower in the siYAP1 agomir group compared with the agomir control group, as evaluated by immunohistochemistry and qRT-PCR (Fig. 1k, l).
YAP1 is upregulated, associated with poor prognosis, and promotes proliferation, invasion, and migration of CRC. a YAP1 was highly expressed in CRC samples in the GSE20916 dataset. b, c YAP1 showed higher levels in CRC samples compared with the corresponding adjacent normal colorectal specimens as assessed by qRT-PCR and immunohistochemistry. d Higher YAP1 expression was associated with poorer OS. e–g YAP1 effects on cell proliferation determined after transfection with siYAP1 and the inhibitor negative control, respectively, in HCT116 cells by monoclonal cell test, CCK8 assay, and EdU staining. h Effects of YAP1 on invasion and migration in HCT116 cells after transfection with siYAP1 and the inhibitor negative control, respectively, as assessed by Transwell assay. i Wound-healing assay of HCT116 cells transfected with siYAP1 or the negative control. j In vivo subcutaneous tumors deriving from HCT116 cells in nude mice after injection of RNAi-YAP1 (siYAP1 agomir) or control. After 28 days, the nude mice were euthanized, and subcutaneous tumors were examined macroscopically. Tumor weight reflects the proliferation ability of HCT116 cells in vivo. Results of three representative mice from each group are shown. k, l Immunohistochemistry and qRT-PCR were employed to measure the protein and mRNA expression levels of YAP1 in subcutaneous tumors from nude mice. Three independent experiments were performed. *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control group by Student’s t-test
YAP1 knockdown downregulates the MALAT1 oncogene in colon cancer cells
Although YAP1 was confirmed as an oncogene in CRC, the underlying mechanism remains undefined. First, gene microarray analysis (GSE92335) [25] revealed differential expression of 11 lncRNAs in HCT116 cells treated with siYAP1 or negative control (Fig. 2a). Similarly, MALAT1 was up-regulated in CRC tissues compared with normal colorectal tissues by analyzing the GSE9348 microarray dataset (Fig. 2b). In addition, MALAT1 was highly expressed in 83 CRC samples, as assessed by qRT-PCR (p < 0.001; Fig. 2c). Moreover, correlation analysis showed that YAP1 expression was closely related to MALAT1 in the GSE14095 dataset, GEPIA database, and 83 CRC samples assessed here (all p < 0.01; Fig. 2d, e). Furthermore, survival analysis indicated that higher expression of MALAT1 was associated with poor OS in all 83 CRC cases (p < 0.05; Fig. 2f); however, the cell localization and biological function of MALAT1 in CRC are largely unknown. MALAT1 was primarily localized in the cytoplasm of colon cancer cells, as estimated by cell fractionation assays (figure S1b). The results for MALAT1 depletion efficiency in vivo and in vitro are shown in figure S4c-d. Moreover, MALAT1 knockdown suppressed invasion and migration in HCT116 cells (figure S1c, d). These results indicated that MALAT1 played an oncogenic role in CRC and might be positively regulated by YAP1.
YAP1 silencing downregulates the oncogene MALAT1 in colon cancer cells. a Heat-map (from GSE92335) showing differential expression of lncRNAs in control (Ctl; negative control) and YAP1-silenced (siYAP1) HCT116 cells. Increasing red color indicates increasing signal strength, whereas weak signal strength is represented by increasing blue color. b Analysis of the GSE14095 dataset showed that MALAT1 expression was positively associated with YAP1 mRNA expression. c MALAT1 was highly expressed in CRC samples compared with corresponding adjacent normal colorectal samples as assessed by qRT-PCR. d, e Correlation analysis of GSE14095 dataset and 83 CRC cases demonstrated that MALAT1 was closely related to YAP1 mRNA expression. f High MALAT1 expression was associated with poor OS. Three independent experiments were performed. *p < 0.05, ***p < 0.001 compared to the control group, by Student’s t-test
YAP1 regulates colon cancer cell proliferation and metastasis through MALAT1
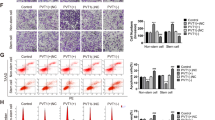
Since MALAT1 expression is regulated by YAP1 interaction with TCF4/β-catenin, whether YAP1 completely or partially acts via MALAT1 is yet to be determined. As shown in Fig. 3, YAP1 overexpression promoted cell migration in HCT116 cells, whereas MALAT1 knockdown reversed the effects of YAP1 overexpression partially, in wound-healing (Fig. 3a) and Transwell migration (Fig. 3b) assays. Next, we established a subcutaneous xenograft model by injection with HCT116 cells stably transfected with control, YAP1, RNAi control, or YAP1 + RNAi-MALAT1. The results showed that YAP1 overexpression promoted the growth of in vivo subcutaneous colon tumors compared with the control group. In addition, RNAi-MALAT1 partially reversed the effect of YAP1 overexpression in promoting in vivo tumor growth (Fig. 3c, d). Moreover, YAP1 overexpression increased in vivo MALAT1 expression in subcutaneous colon tumors, while RNAi-MALAT1 reduced this increasing trend (Fig. 3e). However, RNAi-MALAT1 did not affect YAP1 expression in vivo (Fig. 3f). Overall, the YAP1–MALAT1 axis regulated biological activities in colon cancer cells.
YAP1-induced MALAT1 modulates the biological function of colon cancer cells in vitro and in vivo. a Wound-healing assay of HCT116 cells transfected with pEGFP-YAP1, pEGFP-YAP1 + MALAT1, or vector. b The effect of the YAP1–MALAT1 axis on migration was assessed in HCT116 cells after transfection with pEGFP-YAP1, pEGFP-YAP1 + MALAT1, or vector, by Transwell assay. c, d In vivo subcutaneous tumor xenografts stably transfected HCT116 cells in nude mice after injection of control, YAP1, RNAi control, or YAP1 + RNAi-MALAT1. After 28 days, the nude mice were sacrificed and the subcutaneous tumors were assessed macroscopically. Tumor weight reflected the proliferation ability of HCT116 cells in vivo. Four representative mice from each group are shown. e MALAT1 expression in various groups with in vivo subcutaneous tumors, as assessed by qRT-PCR. f Immunohistochemical detection of YAP1 protein levels in subcutaneous tumors from nude mice. Three independent experiments were performed. *p < 0.05, **p < 0.01 compared to the control group, by Student’s t-test
YAP1 interactions with TCF4 and β-catenin regulate MALAT1 expression in colon cancer cells
To further explore the mechanism by which YAP1 regulates MALAT1, bioinformatics analyses predicted that the MALAT1 promoter had three binding sequences for the transcription factor TCF4 (JASPAR, ifti.org, and UCSC). Interestingly, TCF4 and β-catenin were positively regulated by YAP1 in HCT116 cells as evaluated by qRT-PCR (Fig. 4a) and Western blotting (Fig. 4b). In addition, both TCF4 and β-catenin interacted with the YAP1 protein, as observed by IP (Fig. 4c). Luciferase activity assays indicated that gene knockdown of YAP1, TCF4, and β-catenin inhibited MALAT1 promoter activity in HCT116 and SW480 cells (Fig. 4d), and downregulated MALAT1 in HCT116 cells (Fig. 4e). Further bioinformatics analysis (JASPAR) showed that TCF4 might have a DNA binding motif (Fig. 4f). Next, we analyzed the ChIP-seq data of TCF4 in HCT116 cells from ENCODE. The results showed high signals of TCF4 (TCF7L2) in the MALAT1 promoter region, which suggested a binding potential between TCF4 and the MALAT1 promoter (Figure S3). The interactions among these three genes prompted us to identify the binding site of the MALAT promoter affected. Thus, we constructed luciferase reporters for different predicted binding-sites and observed that the suppression of YAP1, TCF4, or β-catenin reduced the activities of Luc-MAL-pro and Luc-MAL-pro-#1, both containing the first WT binding site away from the start site of MALAT1 transcription. Moreover, Luc-MAL-pro-#2 and Luc-MAL-pro-#3 did not contain the second and third WT binding sites, respectively (Fig. 4g–i). Furthermore, YAP1, TCF4, or β-catenin knockdown affected the activity of MALAT1 promoter containing only the first WT- and non-mutant (MT) binding sites (Fig. 4j). These findings suggested that YAP1 and TCF4 were transcription co-activator and transcription factor, respectively. Then, qPCR-ChIP assays were carried out in HCT116 and SW480 cells. The results showed that anti-TCF4, anti-YAP1, and anti-β-catenin pulled-down the predicted binding-site sequence of the MALAT1 promoter in HCT116 and SW480 cells (Fig. 4k). These findings suggested that YAP1 interacted with TCF4 and β-catenin, and formed a compound regulating MALAT1 in CRC.
YAP1 in combination with TCF4 and β-catenin regulates MALAT1 expression in colon cancer cells. a YAP1, TCF4, and β-catenin expression levels in HCT116 cells transfected with pEGFP-c1-YAP1, siYAP1, vector or negative control by qRT-PCR. b TCF4 and β-catenin expression levels in HCT116 cells treated with pEGFP-c1-YAP1, siYAP1, vector or negative control by Western blotting. c IP confirmed YAP1 interactions with TCF4 and β-catenin in HCT116 cells (GAPDH, IgG as a negative control). d Promoter activities in HCT116 and SW480 cells co-transfected with luciferase reporters for MALAT1 and silencing plasmids (siYAP1, siTCF4, siβ-catenin or negative control, as indicated) for 48 h. e The expression levels of MALAT1 in HCT116 cells treated with silencing plasmids (siYAP1, siTCF4, siβ-catenin or negative control, as indicated) for 48 h. f The TCF4 motif predicted from JASPAR matrix models. g–j Promoter activities from HCT116 cells co-transfected with luciferase reporters of differently predicted binding-site plasmids (Luc-MAL-pro, Luc-MAL-pro-#1, Luc-MAL-pro-#2, or Luc-MAL-pro-#3) and treated with the silencing plasmids siYAP1, siTCF4, siβ-catenin, or negative control, were measured by dual luciferase reporter assay. Luc-MAL-pro, containing three WT binding sites; Luc-MAL-pro-#1, Luc-MAL-pro-#2, and Luc-MAL-pro-#3, containing the first, second, and third WT binding sites, respectively, away from the transcription start site of MALAT1; E4F1 was assessed as the negative control. k ChIP assays using anti-TCF4, anti-YAP1, and anti-β-catenin antibodies were performed in HCT116 and SW480 cells. A control IgG was used as the negative control for IP. Semi-qPCR was used to assess ChIP signals. BRP binding region primer, URP unrelated region primer. Three independent experiments were performed. *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control group, by Student’s t-test
MALAT1 sponges the tumor-suppressor miR-126-5p in colon cancer cells
LncRNAs regulate the tumor biological functions in a variety of pathways, including sponging miRNAs. miR-126-5p had the most significant difference in siMALAT1 transfected HCT116 cells (Table S2). Bioinformatics (DIANA TOOLS) predicted that MALAT1 had two complementary binding sequences for miR-126-5p (Fig. 5a). Meanwhile, miR-126-5p was down-regulated by siMALAT1 transfection in HCT116 cells (Figure S4i), as well as in CRC tissues compared with normal colorectal tissues, via analysis of the two microarray datasets GSE83924 and GSE18392 (Fig. 5b, c). In this study, miR-126-5p was lowly expressed in 83 CRC samples compared with the corresponding normal colorectal samples as assessed by qRT-PCR (p < 0.01; Fig. 5d). From stages I–IV, miR-126-5p expression levels decreased gradually (p < 0.05; Fig. 5e). Correlation analysis demonstrated that miR-126-5p expression was negatively associated with MALAT1 (p < 0.05; Fig. 5f). Reduced miR-126-5p expression reflected poorer prognosis in CRC compared with higher miR-126-5p expression (p < 0.01; Fig. 5g). Biologically, miR-126-5p inhibited cell proliferation in colon cancer cells as determined by monoclonal cell test and CCK8 assay (figure S1e, f). Moreover, miR-126-5p expression suppressed cell invasion and migration as assessed by wound healing and Transwell assays, respectively (figure S1g-h).
MALAT1 sponges the tumor-suppressor miR-126-5p in colon cancer cells. a Predicted sequences of miR-126-5p-binding sites within MALAT1, and MALAT1 MT and miR-126-5p MT sequences at the top and bottom, respectively, were used. b, c miR-126-5p was lowly expressed in CRC samples as assessed by the GSE83924 and GSE18392 datasets. d Analysis of miR-126-5p expression levels in colorectal tissues as detected by qRT-PCR. e Analysis of the expression patterns of miR-126-5p in different TNM stages of CRC by qRT-PCR. f Correlation analysis of miR-126-5p and MALAT1 expression levels in CRC tissues by Spearman’s rank correlation coefficient. g Overall survival analysis was carried out to assess the prognostic effect of miR-126-5p in patients with CRC by the Kaplan–Meier method. h HCT116/SW480 cell lysates were incubated with biotin-labeled MALAT1, and miR-126-5p expression levels were determined by qPCR after pull-down. i HCT116/SW480 cell lysates were incubated with biotin-labeled miR-126-5p, and MALAT1 expression was assayed by qPCR after pull-down. j AGO2-RIP was followed by MALAT1 qPCR to assess MALAT1 levels after miR-126-5p overexpression. k Luciferase activity in HCT116 cells co-transfected with miR-126-5p and luciferase reporters containing MALAT1 or mutant transcript. Data were presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. Three independent experiments were performed. *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control group, by Student’s t-test
To validate the direct binding between MALAT1 and miR-126-5p at the physiological levels, an RNA pull-down assay was performed, with a biotin-labeled-specific MALAT1 probe (Fig. 5h). In this experiment, miR-125a-5p was used as a negative control because it has no binding sequence for MALAT1. An inverse pull-down assay evaluated whether miR-126-5p binds MALAT1. HCT116 or SW480 cells were transfected with biotinylated miR-126-5p, and harvested for pull-down assay. MALAT1 was pulled down; however, several site-specific mutations that disrupt the base pairing between MALAT1 and miR-126-5p prevented miR-126-5p from binding to MALAT1 (Fig. 5i). It is known that miRNAs bind their target genes and cause post-transcriptional suppression in an AGO2-dependent manner. To assess whether MALAT1 was regulated by miR-126-5p in this manner, we performed anti-AGO2 RIP in HCT116 cells transiently transfected with miR-126-5p. Endogenous MALAT1 was specifically enriched in miR-126-5p-overexpressing cells (Fig. 5j), indicating that it was recruited to AGO2-related RNA-induced silencing complexes, and functionally interacted with miR-126-5p. Next, we established a luciferase construct containing MALAT1 or a mutated form [MALAT1 MT (miR-126-5p)]. Luciferase assay showed that miR-126-5p overexpression significantly inhibited the luciferase activity of MALAT1, with reduced effects on the mutated form of MALAT1 (Fig. 5k).
MALAT1–miR-126-5p regulates VEGFA, SLUG, and TWIST in colon cancer cells
Since miR-126-5p was confirmed to be sponged by MALAT1, we next assessed its downstream cascade. MicroRNAs regulate biological functions primarily by inhibiting the expression of target genes. As shown above, miR-126-5p was downregulated and exerted a tumor suppressor function in CRC. Therefore, bioinformatics tools (MIRDB, microRNA.org, and TargetScan.org) were used to predict that miR-126-5p potentially interacted with the 3′UTR regions of SLUG, TWIST, and VEGFA mRNAs through complementary sequences (Fig. 6a). The mRNA expression levels of SLUG, TWIST, and VEGFA, detected by qRT-PCR, were increased (Figure S2a) and negatively correlated with miR-126-5p expression in 83 cases of CRC (Fig. 6b–d). VEGFA, SLUG, and TWIST expression levels were positively associated with MALAT1 expression in the 83 above cases of CRC, GEO data (GSE10495), and GEPIA database, respectively (Fig. 6e–g, S2b, d). In addition, YAP1 expression showed positive associations with VEGFA, SLUG, and TWIST expression levels in the GEO database (GSE10495) and GEPIA database (Figure S2c, d). MiR-126-5p overexpression or MALAT1 knockdown could simultaneously reduce the expression levels of SLUG, TWIST, and VEGFA in colon cancer cells, as assessed by qRT-PCR and Western blotting (Fig. 6h, i). Hence, MALAT1 could regulate SLUG, TWIST, and VEGFA by sponging miR-126-5p in order to exert its biological function in CRC. To further evaluate the ability of miR-126-5p in targeting SLUG, TWIST, and VEGFA, WT and MT plasmids of psiCHECK™-2-SLUG 3′UTR-Luc, psiCHECK™-2-VEGFA XII 3′UTR-Luc, and psiCHECK™-2-TWIST 3′UTR-Luc were constructed. As shown in Fig. 6j, miR-126-5p directly targeted SLUG, TWIST, and VEGFA in a dual luciferase reporter assay system, thereby exhibiting a tumor suppressor role in CRC.
MALAT1–miR-126-5p regulates VEGFA, SLUG, and TWIST expression in colon cancer cells. a Predicted sequences of miR-126-5p binding sites within the 3′-UTRs of VEGFA, SLUG, and TWIST, and sequences of VEGFA, SLUG, and TWIST 3′-UTR mutants (Mut) used in this study (top). b–d Correlation analysis of miR-126-5p and predicted target gene (VEGFA, SLUG, and TWIST) expression levels in CRC tissues, assessed by Spearman’s rank correlation. e–g Correlation analysis of MALAT1 and expression levels of predicted target gene (VEGFA, SLUG, and TWIST) in CRC tissues, assessed by Spearman’s rank correlation. h, i Analysis of miR-126-5p and MALAT1 regulatory effects on VEGFA, SLUG, and TWIST in HCT116 cells by qRT-PCR and immunoblot. j Luciferase activity in HCT116 cells co-transfected with miR-126-5p mimics and luciferase reporters containing the indicated 3′-UTR-driven reporter constructs or mutant transcripts. Data were presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. Three independent experiments were performed. *p < 0.05, **p < 0.01 compared to the control group, by Student’s t-test
The MALAT1–miR-126-5p–VEGFA/SLUG/TWIST axis regulates the biological function of colon cancer cells
The above results demonstrated that MALAT1 directly bound to miR-126-5p and suppressed its activity, which in turn, targeted VEGFA, SLUG, and TWIST. Knockdown of MALAT1 downregulated VEGFA, SLUG, and TWIST in HCT116 cells (Fig. 7a). In rescue experiments, miR-126-5p was inhibited in MALAT1-knockdown HCT116 cells. Inhibition of miR-126-5p restored the decreased levels of VEGFA, SLUG, and TWIST (Fig. 7a). Next, we hypothesized that MALAT1 controls VEGFA, SLUG, and TWIST in a miR-126-5p-dependent manner. MALAT1 over-expression resulted in increased levels of VEGFA, SLUG, and TWIST protein levels, whereas VEGFA, SLUG, and TWIST expression levels in the MALAT1-mut (miR-126-5p) group were similar to control values (Fig. 7b). In addition, the protein expression levels of VEGFA, SLUG, and TWIST were altered by the YAP1–MALAT1–miR-126-5p axis as assessed by Western blotting (Fig. 7c). Subsequently, we assessed the physiological function of the MALAT1–miR-126-5p axis in colon cancer cells. MALAT1 over-expression promoted the migration of HCT116 cells, while migration and angiogenesis in the MALAT1-mut (miR-126-5p) group were similar to those of the control group (Fig. 7d, e). Conversely, MALAT1 knockdown reduced migration and angiogenesis in HCT116 cells (Fig. 7f, g). Similarly, in rescue experiments, miR-126-5p inhibition in MALAT1-downregulated HCT116 cells overcame the declined migration and angiogenesis (Fig. 7f, g).
The MALAT1–miR-126-5p–VEGFA/SLUG/TWIST axis modulates the biological function of colon cancer cells in vitro and in vivo. a Western blot analysis of VEGFA, SLUG, and TWIST protein expression levels after MALAT1 knock-down in HCT116 cells. b Western blot analysis of VEGFA, SLUG, and TWIST expression levels after MALAT1 overexpression in HCT116 cells. c Effects of the YAP1–MALAT1–miR-126-5p axis on YAP1, VEGFA, SLUG, and TWIST protein expression levels in HCT116 cells, as assessed by Western blot. d Migration in Transwell assays after MALAT1 overexpression in HCT116 cells. e Angiogenesis after MALAT1 overexpression in HCT116 cells. f Migration in Transwell assays after MALAT1 knockdown in HCT116 cells. g Angiogenesis after MALAT1 knockdown in HCT116 cells. h Tail vein injection of HCT116 cells transfected control, RNAi-MALAT1, miR-126-5p agomir, and RNAi-MALAT1 + miR-126-5p antagomir into nude mice. After 10 weeks, the mice were euthanized, and metastatic lung nodules were detected macroscopically. i The ratio of weight of lung tissue to weight of whole body reflected the metastasis ability of HCT116 cells in vivo. Results in four representative mice from each group are shown. j Analysis of miR-126-5p expression levels in metastatic lung nodules from nude mice treated with RNAi-MALAT1, miR-126-5p agomir, miR-126-5p antagomir or agomir control, as assessed by qRT-PCR. k Expression of VEGFA, SLUG, and TWIST in metastatic lung nodules of nude mice treated with miR-126-5p agomir, siMALAT1 agomir, or agomir control, determined by qRT-PCR. l Immunohistochemistry for assessing protein levels of VEGFA, SLUG, and TWIST in subcutaneous tumors from nude mice. Three independent experiments were performed. *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control group, by Student’s t-test
To determine whether MALAT1 and miR-126-5p affected colon cancer cell metastasis in vivo, HCT116 cells were injected into the tail vein of nude mice. Subsequently, RNAi-MALAT1, miR-126-5p agomir, RNAi-MALAT1 + miR-126-5p antagomir and control groups were injected into the tail vein of these animals twice per week for 10 weeks. These mice were euthanized, and histological observations revealed that the RNAi-MALAT1, miR-126-5p agomir, and RNAi-MALAT1 + miR-126-5p antagomir groups had smaller numbers of macroscopic lung metastases compared with the agomir control group (Fig. 7h). Meanwhile, RNAi-MALAT1, miR-126-5p agomir, and miR-126-5p antagomir groups overtly reduced the ratio of weight of lung tissue to weight of whole body when HCT116 cells were injected in vivo (Fig. 7i). In addition, miR-126-5p expression levels in lung metastases from the RNAi-MALAT1, miR-126-5p agomir, and miR-126-5p antagomir groups were higher than control values (Fig. 7j). Furthermore, whether the MALAT1–miR-126-5p axis affected tumorigenesis in a VEGFA/SLUG/TWIST-dependent pattern in vivo was assessed. Surprisingly, both MALAT1 suppression and miR-126-5p over-expression reduced the expression levels of VEGFA, SLUG, and TWIST in lung metastasis in vivo (Fig. 7k, l).
Discussion
At present, CRC is one of the most common malignant cancers [1, 2]. Developing an effective treatment for CRC patients is an indispensable prerequisite. In this study, we found that YAP1 inhibition downregulated MALAT1, which exhibited tumor-promoting effects in colon cancer cells via interaction with TCF4 and β-catenin. Further assessment showed that MALAT1 promoted the biological characteristics of colon cancer cells by sponging miR-126-5p for upregulating its targets VEGFA, SLUG, and TWIST. Meanwhile, YAP1-induced MALAT1 induced EMT and angiogenesis in colon cancer cells.
YAP1 is a potent oncogene upregulated in most cancers. Our previous meta-analysis of YAP1 revealed that both overall and nuclear overexpression levels of YAP1 were intimately associated with adverse OS and disease-free survival (DFS) in several cancers, including CRC [26,27,28]. Currently, YAP1 was proved to promote cell proliferation, invasion, and migration in colorectal cancer [29,30,31]. To explore the underlying regulation of YAP1, we previously knock down the YAP1 expression for the following high-throughput sequencing. Interestingly, we not only found the dysregulated mRNA, but also the most lncRNA as we showed, especially MALAT1. It has been reported that overexpressed MALAT1 was involved in regulating tumor progression [32, 33]. The lncRNA MALAT1 shows a positive effect on cancer tumorigenicity and metastasis by regulating the vasculogenesis-like process and angiogenesis [34]. The loss of MALAT1 could lead to the reduction of metastasis in mammary tumors [35]. There are some studies supporting the current findings, which implied that MALAT1 could be as an oncogene in CRC [36, 37]. MALAT1 was also involved in promoting tumor growth and metastasis in CRC. And these researchers found that MALAT1 could be as a liberator to release oncogene PTBP2 from SFPQ/PTBP2 complex [38]. However, the promotion of YAP1 in CRC by regulating MALAT1 still remains elusive. In our study, we reported that high MALAT1 expression was identified in colorectal cancer tissues and showed an oncogene role of MALAT1 in colon cancer cell. Based on these results, we supposed that YAP1 may positively regulate the MALAT1 to promote the CRC progression.
It is known to us, YAP1 is a transcriptional coactivator. A study reported that β-catenin/YAP/TBX5 transcription complex could enhance the expression of RNA component of mitochondrial RNA processing endoribonuclease (RMRP), which has the function in mitochondrial and ribosomal RNA processing, especially noncoding RNA in colon cancer cells [39]. Previous evidence suggests that lncRNA H19 is aberrantly expressed in osteosarcoma and Hh signaling/Yap1 overexpression could induce H19 expression by gene knockdown [40]. However, the regulation mechanism of YAP1 by control lncRNA is still confused. In this study, we used dual luciferase and ChIP experiment to explore a direct binding between MALAT1 promoter and YAP1 interacting with TCF4 and β-catenin, which provides forceful evidence that YAP1 regulating MALAT1 plays an important role in CRC metastasis. However, the mechanism of regulators targeting MALAT1 in cancer is still to be explored.
Increasing studies showed a ceRNA mechanism regulated by MALAT1 in several malignancies. MALAT1 promotes the development of osteosarcoma by regulating HMGB1 via miR-142-3p and miR-129-5p [41]. Down-regulation of miR-146b-5p by MALAT1 in HCC promotes cancer growth and metastasis [42]. The lncRNA MALAT1 interacts with miR-204 to modulate proliferation, migration, and invasion in human hilar cholangiocarcinoma by targeting CXCR4 [43]. A handful of studies have found the function of miR-126-5p in CRC [44, 45]. It showed that low miR-126-5p is related with metastatic colorectal cancer treated with bevacizumab [46]. Furthermore, miR-126 downregulation promotes cell proliferation, migration, and invasion in CRC and induces cell cycle arrest by targeting IRS-1 [47]. Upstream, few studies showed that miR-126 could be regulated by lncRNA to exert its tumor suppressor role. In lung cancer, miR-126 was sponged by lncRNAPVT1-5, which promoted the cell proliferation [48]. Similarly, miR-126 was inhibited by the lncRNA, HOTAIR to enhance the cisplatin resistance in gastric cancer. In this study, we found that MALAT1 had two miRNA response elements (MREs) of miR-126-5p by bioinformatics analysis. Dual luciferase, RNA-IP, and biotin-RNA pull-down assays showed that MALAT1 potentially sponged miR-126-5p. β-catenin/TCF4 complex in colorectal cancer cells could decrease several of these miRNAs (miR-145, miR-126, miR-30e-3p, and miR-139-5p), which markedly suppressed CRC cell growth in vitro. In our study, we proved that YAP1 bound with β-catenin and TCF4 had a positive effect on MALAT1 promoter. High MALAT1 sponged miR-126-5p to enhance the CRC metastasis. These findings strongly suggested that the presence and onco-role of YAP1-induced MALAT1–miR-126-5p axis in CRC. As we all know, miRNAs suppress the expression of target messenger RNAs (mRNAs) in post-transcriptional gene regulatory pathways and initiate either translational suppression or cleavage [49]. Then, the current findings have proved that miR-126-5p could target three mRNAs, including VEGFA, SLUG, and TWIST in CRC. As shown previously, VEGFA promotes angiogenesis [50], while SLUG and TWIST significantly accelerate the EMT [51].
Take together, we found that the oncogene, YAP1 forming a complex with β-catenin/TCF4 bound to the MALAT1 promoter, which could sponge miR-126-5p to promote VEGFA, SLUG, and TWIST expression. Our findings suggested that YAP1-induced MALAT1–miR-126-5p axis may regulate angiogenesis and the EMT to promote metastasis in CRC (Fig. 8)
Materials and methods
Patient tissue specimens and follow-up
Clinical data were obtained from 83 patients who underwent radical resection for colorectal cancer during January 2010–June 2011 at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). All enrolled patients (16–80 years old) were diagnosed with colorectal adenocarcinoma with no other tumors, had the non-emergency operation treatment, and were undergoing no chemotherapy before operation. The 83 CRC cases were 59.49 ± 8.36 years old, and included 47 males and 36 females. There were 9 stage I, 21 stage II, 38 stage III, and 15 stage IV cases. They included 21 left-sided colon, 16 right-sided colon, and 46 rectal carcinomas. Tumor diameters were <4 cm (n = 21), between 4 and 6 cm (n = 44), and >6 cm (n = 18). There were 23 ulcerating, 50 timorous, and 10 infiltrating carcinomas, with 15 distant and 68 non-distant metastasis cases. The patients were divided into high (n = 59) and low YAP1 (n = 24) expression groups based on the median of YAP1 expression in CRC tissues. Then, the associations of malignant phenotype and prognosis with YAP1 expression in these two groups were analyzed. The patients underwent radical surgery for CRC and the malignancy was confirmed by histopathology. According to NCCN Guidelines, the patients were classified as stage II disease and above and administered postoperative chemotherapy. Adjacent normal tissue samples were obtained from normal colorectal tissues localized 5 cm distal to the tumors. None of the patients had a history of radiotherapy or chemotherapy before surgery. Follow-up was performed monthly for the first 6 months, then quarterly until 2 years, every 6 months until 5 years, and once a year thereafter. For follow-up, telephonic interviews and medical examination were performed for 5 years after discharge post-systematic treatment, until June 2016. Survival was calculated by the month. OS was defined as the time from tumor excision to death by any cause. The protocols were approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans; all patients provided written informed consent.
In situ hybridization and immunohistochemistry
In situ hybridization was performed to detect MALAT1 expression in tissue specimens with three 30-base nucleotide probes from different MALAT1 regions. The MALAT1 probes were: 5′-TTGAACTCGCTTTCCATGGCGATTTGCCTTGTGAG-3′, 5′-CTTTAATCTTCCTTCAAAAGGTGGTAAACTATACC-3′, and 5′-TTAAACCGAAGGTGATTAAAAGACCTTGAAATCCA-3′, and synthesized and labeled with DIG-dUTP at the 3′ end by Boster (Wuhan, China; No. 01533TW424056050DL). In situ hybridization was performed as described previously [52]. The semi-quantitative scoring criterion for in situ hybridization was used, wherein both the staining intensity and the number of positive cells were recorded. Protein expression levels of YAP1 were determined by the ElivisionTM two-step immunohistochemical method in normal colorectal and colorectal cancer tissue samples (supplementary material).
Cell culture and transfection
HEK293 and HCT116 cells were cultured in DMEM containing 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), and NCM460, HT29, CaCO2, SW620, and SW480 cells in RPMI-1640 (Thermo, MA, USA) supplemented with 10% FBS. All cell lines were from Cancer Research Institute, Central South University [17, 53]. The pmirGLO-MAL-Luc wild-type (WT), pmirGLO-MAL-Luc MT(miR-126-5p), psiCHECKTM-2-SLUG-3′-UTR, psiCHECKTM-2-TWIST-3′-UTR, and psiCHECKTM-2-VEGFA-3′-UTR WT plasmids, and the corresponding mutants were purchased from Vigene (Rockville, NZ, USA). The pmirGLO-MALAT1 promoter reporter (−2000 to +500 region of the MALAT1 promoter) plasmid and the corresponding mutant were also purchased from Vigene. The pcDNA-YAP1, pcDNA-MALAT1 (functional region) (MALAT-1: lncRNA and its vital 3′ end functional motif in colorectal cancer metastasis), and pcDNA-MALAT1-mut (miR-126-5p) plasmids were obtained from GenePharma (Shanghai, China). The pLVX-TRE3G-ZsGreen1/YAP1 plasmid was obtained from Vigene. The three plasmid groups were mixed with pLVX-TRE3G-ZsGreen1/YAP1, and incubated with HCT116 cells for stable transfection. After 10 days, puromycin-resistant cell pools were established.
MicroRNA (miRNA) mimics and inhibitors, and siRNAs were purchased from Ribobio (Guangzhou, China). The sequences of siRNA are shown in Table S4. The best siYAP1 and siMALAT1 variants were selected for transfection experiments; transfection was performed with Lipofectamine 3000 (Thermo, MA, USA) according to the manufacturer’s instructions.
Gene microarray analysis
HCT116 cells were transfected with scramble siRNA (siCtrl) or siRNA targeting YAP1 (siYAP1) or MALAT1 (siMALAT1) for 36 h. Total RNA was extracted and assessed with Affymetrix Human Genome U133 Plus 2.0 Array (Thermo, MA, USA). The data were deposited in NCBI’s Gene Expression Omnibus (#GSE92335). Gene set enrichment analysis (GSEA) was used to evaluate expression differences in gene sets between the two groups.
Data mining and analysis
Three independent cohorts of primary colorectal cancer GEO data and their correlated clinical data, GSE22598 [54], GSE9348 [55], GSE18392 [56], GSE14095 [57], GSE20916 [58], and GSE83924 [59] were downloaded from the GEO database.
Western blotting
Cell lysis, electrophoresis, and target protein visualization were performed as described previously [60]. See supplementary material for details.
Quantitative RT-PCR (qRT-PCR) and luciferase activity assay
Total RNA was isolated with TRIzol reagent (Thermo, MA, USA) as described previously [61]. The primer sequences are shown in Table S3. Luciferase activity assays were performed with Dual-Luciferase Reporter Assay System (Promega, Beijing, China). Three promoter sequences of MALAT1 are shown in Table S5. See supplementary material for details.
Cell proliferation, invasion, and metastasis assays
Cell proliferation was assayed with CCK-8 (Dojin Laboratories, Tokyo, Japan) and Click-iT® EdU Microplate Assay Kit (Ribobio, Guangzhou, China), according to the manufacturer’s instructions. Wounds were generated with 10 μL pipette tips. Matrigel invasion assay was performed as described previously [62]. See supplementary material for details.
Subcellular fractionation
To determine the cellular localization of MALAT1 in colon cells, cytoplasmic and nuclear fractions were collected according to the manufacturer’s instructions using PARIS Kit (Life Technologies, MA, USA). Data analysis was performed as described previously [63].
Immunoprecipitation (IP) and chromatin immunoprecipitation (ChIP)
Cell lysates were incubated with 2 μg antibody for 16 h at 4 °C. Then, protein A/G Sepharose beads were added to the mixture and incubated for 4 h at 4 °C. Subsequently, the generated immuno-complexes were washed with lysis buffer four times and separated by SDS-PAGE.
ChIP assays were performed as described previously [64]. Formaldehyde-fixed cells were immunoprecipitated overnight with anti-TCF4 antibody (Santa Cruz, CA, USA) or rabbit IgG, and the associated genomic DNA was assessed by PCR and agarose gel electrophoresis. PCR was performed with MALAT1 promoter-specific primers that amplified the YAP1/TCF4/β-catenin-binding regions. The primers were MALAT1 Forward 5′-AGAACAGTTTCACCAGCGTCAAT-3′ and Reverse: 5′-TCCCCACCTGCCCACATAAAG-3′.
RNA-binding protein immunoprecipitation (RIP)
RIP assays were performed with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA) following the manufacturer’s protocol. HCT116 cells were transfected with miR-126-5p control or miR-125a-5p mimic for 48 h, followed by lysis with the RIP lysis buffer. Then, 100 μL of cell extract was incubated with the RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibodies (Abcam, Cambridge, UK) or the negative control (Millipore, MA, USA). After antibody recovery by the protein A/G beads, qRT-PCR was performed to assess the expression levels of MALAT1 in the precipitates. lgG was used to normalize RNA-IPs. The RIP-qPCR Data Analysis (ΔΔCt method) was performed as described previously [65, 66]. Normalize each RIP RNA fractions’ Ct value to the input RNA fraction Ct value for the same qPCR Assay (ΔCt) to account for RNA sample preparation differences.
RNA pull-down assay
RNA pull-down was performed as described previously [67]. SW480 and HCT116 cells were transfected with biotinylated miRNA (40 nmol/L) (Ribobio, Guangzhou, China). After 48 h, the cells were harvested, and lysates were incubated with M-280 streptavidin magnetic beads (Sigma, Shanghai, China). The bound RNAs were purified with TRIzol reagent, and MALAT1 mRNA levels were evaluated by qPCR.
The biotinylated DNA probe complementary to MALAT1 was synthesized (GenePharma, Shanghai, China). SW480 and HCT116 cells were overexpressed with MALAT1, MALAT1-mut (miR-126-5p), or LINC00152 containing plasmids. Then, cell lysates were prepared after 48 h and incubated with the biotinylated probe and M-280 streptavidin magnetic beads (Sigma, Shanghai, China). The miR-126-5p levels were analyzed by qPCR; miR-125a-5p was used as a negative control since it has no binding sequence for MALAT1.
Tube formation assay
Tube formation assay was performed as described previously [68]. In these assays, 96-well plates were coated with 50 μL Matrigel. Then, 2 × 104 HUVEC were co-incubated with 200 μL of supernatants from each group for 10 min, followed by incubation at 37 °C for 6–8 h. Images were acquired under a microscope (Nikon, Tokyo, Japan). Antiangiogenic activity was quantified by measuring the length of the tube walls formed between discrete endothelial cells in each well relative to the control.
Subcutaneous xenotransplantation model
Animal care and euthanasia were approved by the Institutional Animal Care and Use Committee of Zhengzhou University (Changsha, China). According to the completely randomized design using random comparison table, 5-week-old male nude athymic BALB/c nu/nu mice (Slack, Changsha, China) were randomly divided into eight groups: siRNA control, RNAi-YAP1, control (pLVX-TRE3G-ZsGreen1), YAP1 (pLVX-TRE3G-ZsGreen1/YAP1), YAP1 + RNAi-MALAT1, siRNA control, RNAi-MALAT1, miR-126-5p agomir and RNAi-MALAT1 + miR-126-5p antagomir groups, for examining tumorigenicity. There are 3 mice in each group in the experiment of RNAi-YAP1, 4 in the experiment of YAP1 + RNAi-MALAT1, and 4 in the experiment of RNAi-MALAT1 + miR-126-5p antagomir. Logarithmic phase HCT116 cells (4 × 105/100 μL) were inoculated subcutaneously into the dorsal flank and Logarithmic phase HCT116 cells (3 × 106/100 μL) were injected into the tail vein of nude mice. The corresponding groups of nude mice were treated with RNAi-YAP1, RNAi-MALAT1 or control (150 μL, 200 nM), miR-126-5p antagomir and miR-126-5p agomir or control agomir (150 μL, 200 nM) by injection, respectively. The mice inoculated subcutaneously into the dorsal flank were treated with these siRNAs every 3 days for the first 9 days to maintain the siRNA effects for 28 days. The mice injected into the tail vein were treated with these siRNAs every 3 days to maintain the siRNA effects for 10 weeks. The tumor volume was evaluated by the following formula: volume = (width + length)/2 × width × length × 0.5236, and tumor growth curves were generated. All animals were sacrificed 32 days after inoculation, and the tumors were excised, weighed, fixed, and paraffin embedded. A class of long-acting siRNAs (Ribobio Co., Guangzhou, China), agomir, was used as long-acting siRNA because it embeds specific chemical modifications to protect the siRNAs against the immune system in vivo.
Statistical analysis
All statistical analyses were carried out with SPSS version 18.0 (MT, USA) and the GraphPad Prism 5.0 software (CA, USA). Data are mean ± SEM. Group pairs were compared by Student’s t-test. The Pearson’s coefficient was used to assess the correlations between variables. Survival data were obtained by the Kaplan–Meier method, with significance assessed by the log-rank test. The associations of YAP1 expression and clinicopathologic variables were assessed by the Chi-square test or Fisher’s exact test. Median YAP1, MALAT1, and miR-126-5P expression levels were cutoff points for determining high and low expression. p < 0.05 was considered statistically significant.
Change history
28 July 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41388-023-02794-4
References
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–93.
Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Bottomley A. Metastatic colorectal cancer: treatment challenges and quality of life. Lancet. 2002;359:1537–8.
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484.
Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89.
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Clin Gastroenterol Hepatol. 2015;75:6890.
Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6.
Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23:1167.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ, Hsu CW, et al. Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer. Mol Cancer. 2018;17:72.
Matsumura K, Kawasaki Y, Miyamoto M, Kamoshida Y, Nakamura J, Negishi L, et al. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2016;36:1191–9.
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–161.
Ma Y, Yang Y, Feng W, Moyer MP, Wei Q, Peng Z, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–504.
Setonrogers S. Oncogenes: all eyes on YAP1. Nat Rev Cancer. 2014;14:514–5.
Sun D, Li X, He Y, Li W, Wang Y, Wang H, et al. YAP1 enhances cell proliferation, migration, and invasion of gastric cancer in vitro and in vivo. Oncotarget. 2016;7:81062–76.
Ou C, Sun Z, Li X, Ren W, Qin Z, Zhang X, et al. miR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63.
Lu T, Li Z, Yang Y, Ji W, Yu Y, Niu X, et al. The Hippo/YAP1 pathway interacts with FGFR1 signaling to maintain stemness in lung cancer. Cancer Lett. 2018;423:36–46.
Moroishi T, Hansen CG, Guan K-L. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–9.
Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–84.
Zhou Y, Shan T, Ding W, Hua Z, Shen Y, Lu Z, et al. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo-YAP signaling. J Cell Physiol. 2017;233:5805–14.
Di CS, Zhang F, Sanchomedina A, Li S, Aguilo F, Sun Y, et al. RBM-AS1 is critical for self-renewal of colon cancer stem-like cells. Cancer Res. 2016;76:5615–27.
Li S, Yu Z, Chen SS, Li F, Lei CY, Chen XX, et al. The YAP1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding RNA. Urol Oncol. 2015;33:427.e1–10.
Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S, et al. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112:4235.
Ou C, Sun Z, Li X, Ren W, Qin Z, Zhang X, et al. miR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53.
Sun Z, Xu R, Li X, Ren W, Ou C, Wang Q, et al. Prognostic value of Yes-associated protein 1 (YAP1) in Various cancers: a meta-analysis. PLoS ONE 2015;10:e0135119.
Lee KW, Lee SS, Kim SB, Sohn BH, Lee HS, Jnag HJ, et al. Significant association of oncogene YAP1 with poor prognosis and cetuximab resistance in colorectal cancer patients. Clin Cancer Res. 2015;21:357–64.
Li W, Cao Y, Xu J, Wang Y, Li W, Wang Q, et al. YAP transcriptionally regulates COX-2 expression and GCCSysm-4 (G-4), a dual YAP/COX-2 inhibitor, overcomes drug resistance in colorectal cancer. J Exp Clin Cancer Res. 2017;36:144.
Wang Z, Liu P, Zhou X, Wang T, Feng X, Sun YP, et al. Endothelin promotes colorectal tumorigenesis by activating YAP/TAZ. Cancer Res. 2017;77:2413–23.
Ma X, Zhang H, Xue X, Shah YM. Hypoxia-inducible factor 2α (HIF-2α) promotes colon cancer growth by potentiating Yes-associated protein 1 (YAP1) activity. J Biol Chem. 2017;292:17046–56.
Sun M, Song H, Wang S, Zhang C, Zheng L, Chen F, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol. 2017;10:79.
Na KL, Lee JH, Ivan C, Hui L, Zhang X, Chan HP, et al. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer. 2017;17:46.
Li Q, Chao Z, Chen R, Xiong H, Qiu F, Liu S, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016;383:28–40.
Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31.
Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51.
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2013;7:3174.
Li P, Zhang X, Wang H, Wang L, Liu T, Du L, et al. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739–51.
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736.
Park J, Jeong S. Wnt activated β-catenin and YAP proteins enhance the expression of non-coding RNA component of RNase MRP in colon cancer cells. Oncotarget. 2015;6:34658–68.
Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–66.
Liu K, Huang J, Ni J, Song D, Ding M, Wang J, et al. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16:578.
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, et al. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget. 2017;8:28683–95.
Tan X, Huang Z, Li X. Long non-coding RNA MALAT1 interacted with miR-204 to modulate human hilar cholangiocarcinoma proliferation, migration and invasion by targeting CXCR4. J Cell Biochem. 2017;118:3643–53.
Almeida AL, Bernardes MV, Feitosa MR, Peria FM, Tirapelli DP, Rocha JJ, et al. Serological under expression of microRNA-21, microRNA-34a and microRNA-126 in colorectal cancer. Acta Cir Bras. 2016;31:13.
Ebrahimi F, Gopalan V, Wahab R, Lu CT, Anthony SR, Lam AK. Deregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significance. Exp Cell Res. 2015;339:333–41.
Fiala O, Pitule P, Hosek P, Liska V, Sorejs O, Bruha J, et al. The association of miR-126-3p, miR-126-5p and miR-664-3p expression profiles with outcomes of patients with metastatic colorectal cancer treated with bevacizumab. Tumour Biol. 2017;39:1010428317709283.
Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan WK, et al. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS ONE 2013;8:e81203.
Li H, Chen S, Liu J, Guo X, Xiang X, Dong T, et al. Long non-coding RNA PVT1-5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem Biophys Res Commun. 2018;495:2350–5.
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36.
Ghosh A, Dasgupta D, Roychoudhury S, Kumar D, Gorain M, Butti R, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8:e2706.
Yamada N, Sugai T, Eizuka M, Tsuchida K, Sugimoto R, Mue Y, et al. Tumor budding at the invasive front of colorectal cancer may not be associated with the epithelial mesenchymal transition. Hum Pathol. 2016;60:151.
Zeng ZY, Zhou YH, Zhang WL, Xiong W, Fan SQ, Li XL, et al. Gene expression profiling of nasopharyngeal carcinoma reveals the abnormally regulated Wnt signaling pathway. Hum Pathol. 2007;38:120.
Ren W, Sun Z, Zeng Q, Shuang H, Zhang Q, Jiang L. Aberrant expression of CUL4A is associated with IL-6/STAT3 activation in colorectal cancer progression. Arch Med Res. 2016;47:214–22.
Okazaki S, Ishikawa T, Iida S, Ishiguro M, Kobayashi H, Higuchi T, et al. Clinical significance of UNC5B expression in colorectal cancer. Int J Oncol. 2012;40:209–16.
Yi H, Downey T, Kong WE, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90.
Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401.
Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, et al. Gene expression signature and response to the use of leucovorin, fluorouracil and oxaliplatin in colorectal cancer patients. Clin Transl Oncol. 2011;13:419–25.
Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE 2010;5:e13091.
Nagy ZB, Wichmann B, Kalmár A, Galamb O, Barták BK, Spisák S, et al. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin Epigenet. 2017;9:22.
Zhang X, Ai F, Li X, She X, Li N, Tang A, et al. Inflammation-induced S100A8 activates Id3 and promotes colorectal tumorigenesis. Int J Cancer. 2015;137:2803.
Ou C, Sun Z, Zhang H, Xiong W, Ma J, Zhou M, et al. SPLUNC1 reduces the inflammatory response of nasopharyngeal carcinoma cells infected with the EB virus by inhibiting the TLR9/NF-κB pathway. Oncol Rep. 2015;33:2779–88.
Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–37.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409.
Ross J, Bottardi S, Bourgoin V, Wollenschlaeger A, Drobetsky E, Trudel M, et al. Differential requirement of a distal regulatory region for pre-initiation complex formation at globin gene promoters. Nucleic Acids Res. 2009;37:5295–308.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–8.
Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85.
Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596.
Feiyan A, Xuemei Z, Xiayu L, Zailong Q, Qiurong Y, Li T, et al. Up-regulation of matrix metalloproteinases in a mouse model of chemically induced colitis-associated cancer: the role of microRNAs. Oncotarget. 2015;6:5412.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81560385), the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2015211C136), the Medical Scientific and Technological Research Project of Henan Province (201702027), China Postdoctoral Science Foundation (2017M610462), and the Youth Innovation Fund Project of The First Affiliated Hospital of Zhengzhou University (YNQN2017035).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Z., Ou, C., Liu, J. et al. RETRACTED ARTICLE: YAP1-induced MALAT1 promotes epithelial–mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 38, 2627–2644 (2019). https://doi.org/10.1038/s41388-018-0628-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0628-y
This article is cited by
-
KLF15 transcriptionally activates LINC00689 to inhibit colorectal cancer development
Communications Biology (2024)
-
Circ_0091579 Knockdown Inhibited HCC Proliferation and Glutamine Metabolism Through miR-1270/YAP1 Axis
Biochemical Genetics (2024)
-
Mutual interaction of lncRNAs and epigenetics: focusing on cancer
Egyptian Journal of Medical Human Genetics (2023)
-
Polygonum barbatum extract reduces colorectal cancer cell proliferation, migration, invasion, and epithelial–mesenchymal transition via YAP and β-catenin pathway regulation
Scientific Reports (2023)
-
LncRNA MALAT1 promotes growth and metastasis of head and neck squamous cell carcinoma by repressing VHL through a non-canonical function of EZH2
Cell Death & Disease (2023)