Abstract
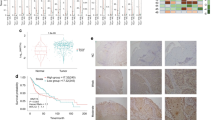
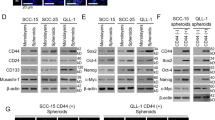
The fibroblast growth factor 19 gene FGF19 has previously been reported to be amplified in several cancer types and encodes for a key autocrine signaler known to promote tumorigenic growth. Thus, it is imperative to understand which cancers are oncogenically addicted to FGF19 amplification as well as the role it serves in these cancer types. We report for the first time high FGF19 amplification in head and neck squamous cell carcinomas (HNSCC), which is associated with increased autocrine secretion of FGF19 and poor patient outcome in HNSCC. FGF19 amplification corresponded with constitutive activation of FGF receptor 4 (FGFR4)-dependent ERK/AKT–p70S6K–S6 signaling activation in HNSCC cells, and addition of human recombinant FGF19 could promote cell proliferation and soft agar colony formation in HNSCC cells with low FGF19 expression through activation of FGFR4 and downstream signaling cascades. In contrast, FGF19 knockout counteracts the observed effects in HNSCC cells carrying high endogenous FGF19, with knockout of FGF19 significantly suppressing tumor growth in an orthotopic mouse model of HNSCC. Collectively, this study demonstrates that FGF19 gene amplification corresponds with an increased dependency upon FGF19/FGFR4 autocrine signaling in HNSCC, revealing a therapeutic target for this cancer type.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma—an update. CA Cancer J Clin. 2015;65:401–21.
Katoh M, Katoh M. Evolutionary conservation of CCND1-ORAOV1-FGF19-FGF4 locus from zebrafish to human. Int J Mol Med. 2003;12:45–50.
Angelin B, Larsson TE, Rudling M. Circulating fibroblast growth factors as metabolic regulators—a critical appraisal. Cell Metab. 2012;16:693–705.
Benoit B, Meugnier E, Castelli M, Chanon S, Vieille-Marchiset A, Durand C, et al. Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nat Med. 2017;23:990.
Glass DJ. What’s so special about FGF19—unique effects reported on skeletal muscle mass and function. Cell Metab. 2017;26:287–8.
Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–82.
Marcelin G, Jo Y-H, Li X, Schwartz GJ, Zhang Y, Dun NJ, et al. Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism. Mol Metab. 2014;3:19–28.
Lang L, Shull A, Teng Y. Interrupting the FGF19-FGFR4 axis to therapeutically disrupt cancer progression. Curr Cancer Drug Targets 2018. https://doi.org/10.2174/1568009618666180319091731
Zhou M, Yang H, Learned RM, Tian H, Ling L. Non-cell-autonomous activation of IL-6/STAT3 signaling mediates FGF19-driven hepatocarcinogenesis. Nat Commun. 2017;8:15433.
Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell. 2011;19:347–58.
Zhao H, Lv F, Liang G, Huang X, Wu G, Zhang W, et al. FGF19 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3β/β-catenin signaling cascade via FGFR4 activation. Oncotarget. 2016;7:13575.
Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7α‐hydroxylase gene expression. Hepatology. 2009;49:297–305.
Gao L, Wang X, Tang Y, Huang S, Hu C-AA, Teng Y. FGF19/FGFR4 signaling contributes to the resistance of hepatocellular carcinoma to sorafenib. J Exp Clin Cancer Res. 2017;36:8–17.
Gao L, Shay C, Lv F, Wang X, Teng Y. Implications of FGF19 on sorafenib-mediated nitric oxide production in hepatocellular carcinoma cells—a short report. Cell Oncol. 2018;41:85–91.
Wan ZY, Tian JS, Tan HWS, Chow AL, Sim AYL, Ban KHK, et al. Mechanistic target of rapamycin complex 1 is an essential mediator of metabolic and mitogenic effects of fibroblast growth factor 19 in hepatoma cells. Hepatology. 2016;64:1289–301.
Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, et al. First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 2015;5:424–37.
Zhao X, Xu F, Dominguez NP, Xiong Y, Xiong Z, Peng H, et al. FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol Carcinog. 2018;57:1616–25.
Schuuring E. The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes—a review. Gene. 1995;159:83–96.
Schwab M. Oncogene amplification in solid tumors. Elsevier. 1999. p. 319-25.
Morrison C, Zanagnolo V, Ramirez N, Cohn DE, Kelbick N, Copeland L, et al. HER-2 is an independent prognostic factor in endometrial cancer: association with outcome in a large cohort of surgically staged patients. J Clin Oncol. 2006;24:2376–85.
Gibcus JH, Menkema L, Mastik MF, Hermsen MA, de Bock GH, van Velthuysen M-LF, et al. Amplicon mapping and expression profiling identify the Fas-associated death domain gene as a new driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Cancer Clin Cancer Res. 2007;13:6257–66.
Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–6.
Tiong KH, Tan BS, Choo HL, Chung FF-L, Hii L-W, Tan SH, et al. Fibroblast growth factor receptor 4 (FGFR4) and fibroblast growth factor 19 (FGF19) autocrine enhance breast cancer cells survival. Oncotarget. 2016;7:57633.
Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–4.
de Mello RA, Gerós S, Alves MP, Moreira F, Avezedo I, Dinis J. Cetuximab plus platinum-based chemotherapy in head and neck squamous cell carcinoma: a retrospective study in a single comprehensive European cancer institution. PLoS ONE. 2014;9:e86697.
Mandic A, Hansson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem. 2003;278:9100–6.
Cardinali M, Pietraszkiewicz H, Ensley JF, Robbins KC. Tyrosine phosphorylation as a marker for aberrantly regulated growth-promoting pathways in cell lines derived from head and neck malignancies. Int J Cancer. 1995;61:98–103.
Yeudall WA, Crawford RY, Ensley J, Robbins K. MTS1/CDK4I is altered in cell lines derived from primary and metastatic oral squamous cell carcinoma. Carcinogenesis. 1994;15:2683–6.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281.
Lang L, Shay C, Xiong Y, Thakkar P, Chemmalakuzhy R, Wang X, et al. Combating head and neck cancer metastases by targeting Src using multifunctional nanoparticle-based saracatinib. J Hematol Oncol. 2018;11:85.
Teng Y, Ren M, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103:1066.
Xie X, Tang S, Cai Y, Pi W, Deng L, Wu G, et al. Suppression of breast cancer metastasis through the inactivation of ADP-ribosylation factor 1. Oncotarget. 2016;7:58111.
Teng Y, Cai Y, Pi W, Gao L, Shay C. Augmentation of the anticancer activity of CYT997 in human prostate cancer by inhibiting Src activity. J Hematol Oncol. 2017;10:118.
Acknowledgements
This work was supported in part by Dental College of Georgia Special Funding Initiative and a grant from the Department of Defense (W81XWH-14-1-0412) (to Y.T.). The authors sincerely thank Dr. Andrew W. Yeudall (Augusta University) for providing HNSCC cells for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Lixia Gao, Liwei Lang
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Gao, L., Lang, L., Zhao, X. et al. FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene 38, 2394–2404 (2019). https://doi.org/10.1038/s41388-018-0591-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0591-7
This article is cited by
-
FGF19 and its analog Aldafermin cooperate with MYC to induce aggressive hepatocarcinogenesis
EMBO Molecular Medicine (2024)
-
FGF19 promotes nasopharyngeal carcinoma progression by inducing angiogenesis via inhibiting TRIM21-mediated ANXA2 ubiquitination
Cellular Oncology (2024)
-
Mediation of PKM2-dependent glycolytic and non-glycolytic pathways by ENO2 in head and neck cancer development
Journal of Experimental & Clinical Cancer Research (2023)
-
CD44 and CD221 directed magnetic cubosomes for the targeted delivery of helenalin to rhabdomyosarcoma cells
Nano Research (2023)
-
First-in-Human Study of INCB062079, a Fibroblast Growth Factor Receptor 4 Inhibitor, in Patients with Advanced Solid Tumors
Targeted Oncology (2023)