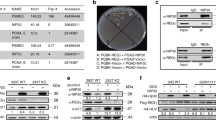
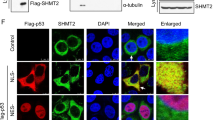
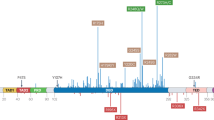
Abstract
Tumor cells with p53 inactivation frequently exhibit chemotherapy resistance, which poses a long-standing challenge to cancer treatment. Here we unveiled a previously unrecognized role of TET2 in mediating p53-loss induced chemotherapy resistance in colon cancer. Deletion of TET2 in p53-null colon cancer cells enhanced DNA damage and restored chemotherapy sensitivity. By taking a two-pronged approach that combined pharmacological inhibition with genetic depletion, we discovered that p53 destabilized TET2 at the protein level by promoting its autophagic degradation. At the molecular level, we further revealed a physical association between TET2 and p53 that facilitated the nucleoplasmic shuttling of TET2, as well as its recruitment to the autophagosome for degradation. Our study has unveiled a functional interplay between TET2 and p53 during anti-cancer therapy. Our findings establish the rationale for targeting TET2 to overcome chemotherapy resistance associated with mutant p53 tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hientz K, Mohr A, Bhakta-Guha D, Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2016;8:8921–46.
Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70.
Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8.
Shetzer Y, Solomon H, Koifman G, Molchadsky A, Horesh S, Rotter V. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis. 2014;35:1196–208.
Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402.
Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–95.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5- methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5.
Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5- hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–13.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5- methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3.
Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43.
Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–9.
Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194.
Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–82.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301.
Tefferi A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Patnaik MM, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–5.
Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38.
Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–9.
Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–7.
Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30:464–74.
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2012;32:663–9.
Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–8.
Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D’Alessio AC, et al. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell. 2015;57:247–60.
An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071.
Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–6.
Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6:278–84.
Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, et al. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell. 2017;65:323–35.
Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–9.
White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10.
Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA. 2010;107:3722–7.
Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–67.
Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol. 2013;65:157–70.
Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33.
Dunkern TR, Wedemeyer I, Baumgartner M, Fritz G, Kaina B. Resistance of p53 knockout cells to doxorubicin is related to reduced formation of DNA strand breaks rather than impaired apoptotic signaling. DNA Repair. 2003;2:49–60.
Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–20.
Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–55.
Kafer GR, Li X, Horii T, Suetake I, Tajima S, Hatada I, et al. 5-hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep. 2016;14:1283–92.
Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73.
Muller PA, Vousden KH.Mutant p53 cancer: new functions and therapeutic opportunities.Cancer Cell. 2014;25:304–17.
Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–83.
Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–8.
Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46.
Jiang D, Wei S, Chen F, Zhang Y, Li J. TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 2017;18:781–96.
Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–9.
Acknowledgements
This work was supported by grants from Cancer Prevention and Research Institute of Texas (RR140053 to YH, to RP170660 to YZ, RR150085 to LH), the Innovation Award from American Heart Association (16IRG27250155 to YH), the John S. Dunn Foundation Collaborative Research Award (to YH), the Center for Translational Environmental Health Research (CTEHR) Seed Grant to YH, the National Institute of Health grants (R01HL134780 to YH, R01GM112003 to YZ), the Welch Foundation (BE-1913 to YZ), the American Cancer Society (RSG-18-043-01-LIB to YH, RSG-16-215-01-TBE to YZ), and by an allocation from the Texas A&M University start- up funds (YH).
Author contributions
YH and YZ conceived, directed and oversaw the project. JZ, JM and W-GD collected CRC samples, p53 mutation analysis and western blotting analysis. JL and DS analyzed the CRC western quantification data. JZ and LG performed the western blotting, doxorubicin / cisplatin treatment, gamma irradiation experiments. JZ and LG performed WST-1, colony-forming assays and quantitative real-time PCR analysis. JZ and ML performed the dot-blot assay. JZ and SF performed immunofluorescent staining. JZ generated majority cell lines used in this study. TP provided cell lines lacking autophagosome components and cell cycle blocking experiments. JG and LH performed the mutational analysis in AML patients. GJ and RD provided colon cancer cell lines. JJ, W-MC, RD and W-GD provided intellectual inputs. YH and YZ wrote the manuscript with all the other authors participating in discussion, data interpretation and manuscript editing.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally to the work: Jixiang Zhang, Peng Tan, Lei Guo
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, J., Tan, P., Guo, L. et al. p53-dependent autophagic degradation of TET2 modulates cancer therapeutic resistance. Oncogene 38, 1905–1919 (2019). https://doi.org/10.1038/s41388-018-0524-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0524-5
This article is cited by
-
TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
Signal Transduction and Targeted Therapy (2023)
-
Clonal hematopoiesis and cardiovascular disease: deciphering interconnections
Basic Research in Cardiology (2022)
-
Correlation of mutational landscape and survival outcome of peripheral T-cell lymphomas
Experimental Hematology & Oncology (2021)
-
The function and regulation of TET2 in innate immunity and inflammation
Protein & Cell (2021)