Abstract
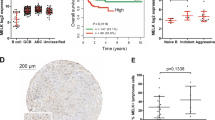
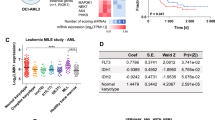
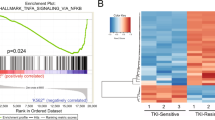
TP53 pathway defects contributed to therapy resistance and adverse clinical outcome in chronic lymphocytic leukemia (CLL), which represents an unmet clinical need with few therapeutic options. Maternal embryonic leucine zipper kinase (MELK) is a novel oncogene, which plays crucial roles in mitotic progression and stem cell maintenance. OTSSP167, an orally administrated inhibitor targeting MELK, is currently in a phase I/II clinical trial in patients with advanced breast cancer and acute myeloid leukemia. Yet, no investigation has been elucidated to date regarding the oncogenic role of MELK and effects of OTSSP167 in chronic lymphocytic leukemia (CLL). Previous studies confirmed MELK inhibition abrogated cancer cell survival via p53 signaling pathway. Thus, we aimed to determine the biological function of MELK and therapeutic potential of OTSSP167 in CLL. Herein, MELK over-expression was observed in CLL cells, and correlated with higher WBC count, advanced stage, elevated LDH, increased β2-MG level, unmutated IGHV, positive ZAP-70, deletion of 17p13 and inferior prognosis of CLL patients. In accordance with functional enrichment analyses in gene expression profiling, CLL cells with depletion or inhibition of MELK exhibited impaired cell proliferation, enhanced fast-onset apoptosis, induced G2/M arrest, attenuated cell chemotaxis and promoted sensitivity to fludarabine and ibrutinib. However, gain-of-function assay showed increased cell proliferation and cell chemotaxis. In addition, OTSSP167 treatment reduced phosphorylation of AKT and ERK1/2. It decreased FoxM1 phosphorylation, expression of FoxM1, cyclin B1 and CDK1, while up-regulating p53 and p21 expression. Taken together, MELK served as a candidate of therapeutic target in CLL. OTSSP167 exhibits potent anti-tumor activities in CLL cells, highlighting a novel molecule-based strategy for leukemic interventions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lu K, Wang X. Therapeutic advancement of chronic lymphocytic leukemia. J Hematol Oncol. 2012;5:55.
Speers C, Zhao SG, Kothari V, Santola A, Liu M, Wilder-Romans K, et al. Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin Cancer Res. 2016;22:5864–75.
Gu C, Banasavadi-Siddegowda YK, Joshi K, Nakamura Y, Kurt H, Gupta S, et al. Tumor-specific activation of the C-JUN/MELK pathway regulates glioma stem cell growth in a p53-dependent manner. Stem Cells. 2013;31:870–81.
Janostiak R, Rauniyar N, Lam TT, Ou J, Zhu LJ, Green MR, et al. MELK promotes melanoma growth by stimulating the NF-κB pathway. Cell Rep. 2017;21:2829–41.
Pickard MR, Green AR, Ellis IO, Caldas C, Hedge VL, Mourtada-Maarabouni M, et al. Dysregulated expression of Fau and MELK is associated with poor prognosis in breast cancer. Breast Cancer Res. 2009;11:R60.
Li S, Li Z, Guo T, Xing XF, Cheng X, Du H, et al. Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget. 2016;7:6266–80.
Du T, Qu Y, Li J, Li H, Su L, Zhou Q, et al. Maternal embryonic leucine zipper kinase enhances gastric cancer progression via the FAK/Paxillin pathway. Mol Cancer. 2014;13:100.
Hiwatashi K, Ueno S, Sakoda M, Iino S, Minami K, Yonemori K, et al. Expression of maternal embryonic leucine zipper kinase (MELK) correlates to malignant potentials in hepatocellular carcinoma. Anticancer Res. 2016;36:5183–8.
Inoue H, Kato T, Olugbile S, Tamura K, Chung S, Miyamoto T, et al. Effective growth-suppressive activity of maternal embryonic leucine-zipper kinase (MELK) inhibitor against small cell lung cancer. Oncotarget. 2016;7:13621–33.
Alachkar H, Mutonga MB, Metzeler KH, Fulton N, Malnassy G, Herold T, et al. Preclinical efficacy of maternal embryonic leucine-zipper kinase (MELK) inhibition in acute myeloid leukemia. Oncotarget. 2014;5:12371–82.
Stefka AT, Park JH,Matsuo Y,Chung S,Nakamura Y,Jakubowiak AJ, et al.Anti-myeloma activity of MELK inhibitor OTS167: effects on drug-resistant myeloma cells and putative myeloma stem cell replenishment of malignant plasmacells. Blood cancer J. 2016;6:e460.
Bolomsky A, Heusschen R, Schlangen K, Stangelberger K, Muller J, Schreiner W, et al. Maternal embryonic leucine zipper kinase is a novel target for proliferation-associated high-risk myeloma. Haematologica. 2018;103:325–35.
Chung S, Suzuki H, Miyamoto T, Takamatsu N, Tatsuguchi A, Ueda K, et al. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3:1629–40.
Chung S, Kijima K, Kudo A, Fujisawa Y, Harada Y, Taira A, et al. Preclinical evaluation of biomarkers associated with antitumor activity of MELK inhibitor. Oncotarget. 2016;7:18171–82.
Ghia P. Ibrutinib holds promise for patients with 17p deletion CLL. Lancet Oncol. 2016;17:1342–3.
Komarova NL, Burger JA, Wodarz D. Evolution of ibrutinib resistance in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci USA. 2014;111:13906–11.
Svirnovski AI, Serhiyenka TF, Kustanovich AM, Khlebko PV, Fedosenko VV, Taras IB, et al. DNA-PK, ATM and MDR proteins inhibitors in overcoming fludarabine resistance in CLL cells. Exp Oncol. 2010;32:258–62.
Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells. 2013;31:1051–63.
Longo PG, Laurenti L, Gobessi S, Petlickovski A, Pelosi M, Chiusolo P, et al. The Akt signaling pathway determines the different proliferative capacity of chronic lymphocytic leukemia B-cells from patients with progressive and stable disease. Leukemia. 2007;21:110–20.
Halasi M, Gartel AL. FOX(M1) news–it is cancer. Mol Cancer Ther. 2013;12:245–54.
Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–33.
Xia H, Kong SN, Chen J, Shi M, Sekar K, Seshachalam VP, et al. MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence. Cancer Lett. 2016;383:85–93.
Kim SH, Joshi K, Ezhilarasan R, Myers TR, Siu J, Gu C, et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem Cell Rep. 2015;4:226–38.
Krishnan A, K D, Babu PSS, Jagadeeshan S, Prasad M, Nair SA. Oncogenic actions of SKP2 involves deregulation of CDK1 Turnover Mediated by FOXM1. J Cell Biochem. 2017;118:797–807.
Motiwala T, Kutay H, Zanesi N, Frissora FW, Mo X, Muthusamy N, et al. PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model. Leukemia. 2015;29:1350–9.
Jurmeister S, Ramos-Montoya A, Sandi C, Pértega-Gomes N, Wadhwa K, Lamb AD et al. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Mol Med. 2018. https://doi.org/10.15252/emmm.201708274.
Wang Y, Lee YM, Baitsch L, Huang A, Xiang Y, Tong H, et al. MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. eLife. 2014;3:e01763.
Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, et al. The target landscape of clinical kinase drugs. Science. 2017. https://doi.org/10.1126/science.aan4368.
Choi S, Ku JL. Resistance of colorectal cancer cells to radiation and 5-FU is associated with MELK expression. Biochem Biophys Res Commun. 2011;412:207–13.
Ciccone M, Ferrajoli A, Keating MJ, Calin GA. SnapShot: chronic lymphocytic leukemia. Cancer Cell. 2014;26:770–e1.
Parikh SA, Shanafelt TD. Prognostic factors and risk stratification in chronic lymphocytic leukemia. Semin Oncol. 2016;43:233–40.
Kohler RS, Kettelhack H, Knipprath-Meszaros AM, Fedier A, Schoetzau A, Jacob F, et al. MELK expression in ovarian cancer correlates with poor outcome and its inhibition by OTSSP167 abrogates proliferation and viability of ovarian cancer cells. Gynecol Oncol. 2017;145:159–66.
Beke L, Kig C, Linders JT,Boens S, Boeckx A,van Heerde E, et al. MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells. Biosci Rep. 2015;35:e00267.
Wiestner A. The role of B-cell receptor inhibitors in the treatment of patients with chronic lymphocytic leukemia. Haematologica. 2015;100:1495–507.
Mertens D, Stilgenbauer S. Ibrutinib-resistant CLL: unwanted and unwonted! Blood. 2017;129:1407–9.
Lenz G. Deciphering Ibrutinib Resistance in Chronic Lymphocytic Leukemia. J Clin Oncol. 2017;35:1451–2.
Kaur V, Swami A. Ibrutinib in CLL: a focus on adverse events, resistance, and novel approaches beyond ibrutinib. Ann Hemato. 2017;96:1175–84.
Davezac N, Baldin V, Blot J, Ducommun B, Tassan JP. Human pEg3 kinase associates with and phosphorylates CDC25B phosphatase: a potential role for pEg3 in cell cycle regulation. Oncogene. 2002;21:7630–41.
Kwok CT, Leung MH, Qin J, Qin Y, Wang J, Lee YL, et al. The Forkhead box transcription factor FOXM1 is required for the maintenance of cell proliferation and protection against oxidative stress in human embryonic stem cells. Stem Cell Res. 2016;16:651–61.
Sullivan C, Liu Y, Shen J, Curtis A, Newman C, Hock JM, et al. Novel interactions between FOXM1 and CDC25A regulate the cell cycle. PLoS ONE. 2012;7:e51277.
Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990–1002.
Cui J, Xia T, Xie D, Gao Y, Jia Z, Wei D, et al. HGF/Met and FOXM1 form a positive feedback loop and render pancreatic cancer cells resistance to Met inhibition and aggressive phenotypes. Oncogene. 2016;35:4708–18.
Liu Y, Chen X, Gu Y, Zhu L, Qian Y, Pei D, et al. FOXM1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and mediates sensitivity to cisplatin in A549 cells via the JNK/mitochondrial pathway. Neoplasma. 2015;62:61–71.
Wang K, Zhu X, Zhang K, Zhu L, Zhou F. FoxM1 inhibition enhances chemosensitivity of docetaxel-resistant A549 cells to docetaxel via activation of JNK/mitochondrial pathway. Acta Biochim Biophys Sin. 2016;48:804–9.
Buchner M, Park E, Geng H, Klemm L, Flach J, Passegué E, et al. Identification of FOXM1 as a therapeutic target in B-cell lineage acute lymphoblastic leukaemia. Nat Commun. 2015;10:6471.
Khan I, Halasi M, Zia MF, Gann P, Gaitonde S, Mahmud N, et al. Nuclear FOXM1 drives chemoresistance in AML. Leukemia. 2017;31:251–5.
Li PP, Feng LL, Chen N, Ge XL, Lv X, Lu K, et al. Metadherin contributes to the pathogenesis of chronic lymphocytic leukemia partially through Wnt/beta-catenin pathway. Med Oncol. 2015;32:479.
Simon M, Mesmar F, Helguero L, Williams C. Genome-wide effects of MELK-inhibitor in triple-negative breast cancer cells indicate context-dependent response with p53 as a key determinant. PLoS ONE. 2017;12:e0172832.
Matsuda T, Kato T, Kiyotani K, Tarhan YE, Saloura V, Chung S, et al. p53-independent p21 induction by MELK inhibition. Oncotarget. 2017;8:57938–47.
Hasegawa K, Ikeda Y, Kunugi Y, Kurosaki A, Imai Y, Kohyama S, et al. Phase I study of multiple epitope peptide vaccination in patients with recurrent or persistent cervical cancer. J Immunother. 2018. https://doi.org/10.1097/CJI.0000000000000214.
Lin A, Giuliano CJ, Sayles NM, Sheltzer JM. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. eLife. 2017;6:e24179.
Ji W, Arnst C, Tipton AR, Bekier ME 2nd, Taylor WR, Yen TJ, et al. OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS ONE. 2016;11:e0153518.
Huang HT, Seo HS, Zhang T, Wang Y, Jiang B, Li Q, et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife. 2017. https://doi.org/10.7554/eLife.26693.
Giuliano CJ, Lin A, Smith JC, Palladino AC, Sheltzer JM. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife. 2018. https://doi.org/10.7554/eLife.32838.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56.
Lu K, Fang XS, Feng LL, Jiang YJ, Zhou XX, Liu X, et al. The STAT3 inhibitor WP1066 reverses the resistance of chronic lymphocytic leukemia cells to histone deacetylase inhibitors induced by interleukin-6. Cancer Lett. 2015;359:250–8.
Zhou X, Fang X, Jiang Y, Geng L, Li X, Li Y, et al. Klotho, an anti-aging gene, acts as a tumor suppressor and inhibitor of IGF-1R signaling in diffuse large B cell lymphoma. J Hematol Oncol. 2017;10:37.
Trojani A, Di Camillo B, Tedeschi A, Lodola M, Montesano S, Ricci F, et al. Gene expression profiling identifies ARSD as a new marker of disease progression and the sphingolipid metabolism as a potential novel metabolism in chronic lymphocytic leukemia. Cancer Biomark. 2011;11:15–28.
Herold T, Mulaw MA, Jurinovic V, Seiler T, Metzeler KH, Dufour A, et al. High expression of MZB1 predicts adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma and is associated with a unique gene expression signature. Leuk Lymphoma. 2013;54:1652–7.
Herold T, Jurinovic V, Metzeler KH, Boulesteix AL, Bergmann M, Seiler T, et al. An eight-gene expression signature for the prediction of survival and time to treatment in chronic lymphocytic leukemia. Leukemia. 2011;25:1639–45.
Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–37.
Kohlmann A, Kipps TJ, Rassenti LZ, Downing JR, Shurtleff SA, Mills KI, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br J Haematol. 2008;142:802–7.
Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K, et al. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood. 2014;124:84–95.
Chuang HY, Rassenti L, Salcedo M, Licon K, Kohlmann A, Haferlach T, et al. Subnetwork-based analysis of chronic lymphocytic leukemia identifies pathways that associate with disease progression. Blood. 2012;120:2639–49.
Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, et al. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73.
Acknowledgements
This study was partly supported by: National Natural Science Foundation (No. 81270598, No. 81473486, No. 81770210), Key Research and Development Program of Shandong Province (No. 2018CXGC1213, No. 2016GSF201029), Natural Science Foundation of Shandong Province (No. ZR2012HZ003), Technology Development Projects of Shandong Province (No. 2014GSF118021), Program of Shandong Medical Leading Talent, and Taishan Scholar Foundation of Shandong Province.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, Y., Zhou, X., Li, Y. et al. Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene 37, 5520–5533 (2018). https://doi.org/10.1038/s41388-018-0333-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0333-x
This article is cited by
-
Discovery of potent maternal embryonic leucine zipper kinase (MELK) inhibitors of novel chemotypes using structure-based pharmacophores
Medicinal Chemistry Research (2023)
-
FOXM1 promotes TGF-β2-induced injury of human lens epithelial cells by up regulating VEGFA expression
Graefe's Archive for Clinical and Experimental Ophthalmology (2023)
-
Targeting metabolic reprogramming in chronic lymphocytic leukemia
Experimental Hematology & Oncology (2022)
-
Competing endogenous RNA networks related to prognosis in chronic lymphocytic leukemia: comprehensive analyses and construction of a novel risk score model
Biomarker Research (2022)
-
Circ_0007031 Silencing Inhibits Cell Proliferation and Induces Cell Apoptosis via Downregulating MELK at a miR-485-3p-Dependent Way in Colorectal Cancer
Biochemical Genetics (2022)