Abstract
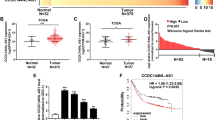
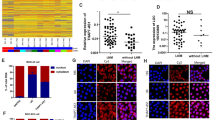
Recently, long noncoding RNAs (lncRNAs) have been reported to play a pivotal role in the occurrence and progression of cancer because of their unique characteristics and have therefore become an active area of cancer research. The object of this study was to screen lncRNAs that are dysregulated in gastric cancer and to investigate their potential functions. Global expression of lncRNAs in gastric cancer and adjacent normal tissues of patients was profiled using a microarray assay. We identified an lncRNA (GALNT5 uaRNA, UTR-associated RNA) that is derived from the 3′-UTR of GALNT5. This lncRNA was transcribed independently of the coding region of GALNT5 and was determined to be markedly upregulated in human gastric carcinoma relative to their corresponding normal gastric tissues by quantitative RT-PCR (qRT-PCR) analysis of tissues from 122 gastric carcinoma patients. The expression of GALNT5 uaRNA was significantly correlated with the TNM stage and with lymph node metastasis. Further results demonstrated that GALNT5 uaRNA facilitated the proliferation and migration of gastric cancer cells in vitro and promoted tumor growth in a mouse model of human gastric cancer. Our results also indicated that GALNT5 uaRNA might function in gastric cancer by binding with HSP90. Further studies indicated that the 5′-end stem-loop motifs of GALNT5 uaRNA promoted the binding of HSP90 and its client proteins, and thus inhibited ubiquitination of the clients. These results expanded our understanding of GALNT5 uaRNA as a new avenue for therapeutic intervention against gastric cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell . 2013;152:1298–307.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63.
Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11.
Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–64.
Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–29.
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–310.
Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35:5770–80.
Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–7.
Mazumder B, Seshadri V, Fox PL. Translational control by the 3’UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–8.
Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, et al. Regulated posttranscriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–50.
Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, et al. Expression of distinct RNAs from 3¢ untranslated regions. Nucleic Acids Res. 2011;39:2393–403.
Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–76.
Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–5.
Rodrigues TC, Fidalgo F, da Costa CM, Ferreira EN, da Cunha IW, Carraro DM, et al. Upregulated genes at 2q24 gains as candidate oncogenes in hepatoblastomas. Future Oncol. 2014;10:2449–57.
Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510.
Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Mol Cell. 2015;58:8–20.
Khurana N, Bhattacharyya S. Hsp90, the concertmaster: tuning transcription. Front Oncol. 2015;5:100.
Tsutsumi S, Neckers L. Extracellular heat shock protein 90: a role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 2007;98:1536–9.
Bishop SC, Burlison JA, Blagg BS. Hsp90: a novel target for the disruption of multiple signaling cascades. Curr Cancer Drug Targets. 2007;7:369–88.
Vahid S, Thaper D, Zoubeidi A. Chaperoning the cancer: the proteostatic functions of the heat shock proteins in cancer. Recent Pat Anticancer Drug Discov. 2017;12:35–47.
Lianos GD, Alexiou GA, Mangano A, Mangano A, Rausei S, Boni L, et al. The role of heat shock proteins in cancer. Cancer Lett. 2015;360:114–8.
Ten Hagen KG, Hagen FK, Balys MM, Beres TM, Van Wuyckhuyse B, Tabak LA. Cloning and expression of a novel, tissue specifically expressed member of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family. J Biol Chem. 1998;273:27749–54.
Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type Oglycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–56.
He H, Shen Z, Zhang H, Wang X, Tang Z, Xu J, et al. Clinical significance of polypeptide Nacetylgalactosaminyl transferase-5 (GalNAc-T5) expression in patients with gastric cancer. Br J Cancer. 2014;110:2021–9.
Kuersten S, Goodwin EB. The power of the 3’ UTR: translational control and development. Nat Rev Genet. 2003;4:626–37.
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7.
Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, et al. Expression of distinct RNAs from 3’ untranslated regions. Nucleic Acids Res. 2011;39:2393–403.
Furuno M, Pan KC, Ninomiya N, Fukuda S, Frith MC, Bult C, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37.
Chen G, Cao P, Goeddel DV. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol Cell. 2002;9:401–10.
Broemer M, Krappmann D, Scheidereit C. Requirement of Hsp90 activity for IkappaB kinase (IKK) biosynthesis and for constitutive and inducible IKK and NF-kappaB activation. Oncogene. 2004;23:5378–86.
Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome independent degradation of IkappaB kinase (IKK). Cell Res. 2006;16:895–901.
Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–66.
Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5.
Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31:4415–27.
Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteom. 2008;7:560–72.
Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40.
Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol Ther. 2015;151:72–86.
Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850–63.
Funding
This work was partially supported by the Natural Science Foundation of China (Grant No. 81572755, 81372200 to J. Shi), CAMS Initiative for Innovative Medicine (CAMS-I2M Grant No. 2016-I2M-1-001to J. Shi).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Guo, H., Zhao, L., Shi, B. et al. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene 37, 4505–4517 (2018). https://doi.org/10.1038/s41388-018-0266-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0266-4
This article is cited by
-
Alternative polyadenylation: methods, mechanism, function, and role in cancer
Journal of Experimental & Clinical Cancer Research (2021)
-
Lnc-GAN1 expression is associated with good survival and suppresses tumor progression by sponging mir-26a-5p to activate PTEN signaling in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2021)
-
Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer
In Vitro Cellular & Developmental Biology - Animal (2019)