Abstract
Tumor stroma has an active role in the initiation, growth, and propagation of many tumor types by secreting growth factors and modulating redox status of the microenvironment. Although PPARβ/δ in fibroblasts was shown to modulate oxidative stress in the wound microenvironment, there has been no evidence of a similar effect in the tumor stroma. Here, we present evidence of oxidative stress modulation by intestinal stromal PPARβ/δ, using a FSPCre-Pparb/d−/− mouse model and validated it with immortalized cell lines. The FSPCre-Pparb/d−/− mice developed fewer intestinal polyps and survived longer when compared with Pparb/dfl/fl mice. The pre-treatment of FSPCre-Pparb/d−/− and Pparb/dfl/fl with antioxidant N-acetyl-cysteine prior DSS-induced tumorigenesis resulted in lower tumor load. Gene expression analyses implicated an altered oxidative stress processes. Indeed, the FSPCre-Pparb/d−/− intestinal tumors have reduced oxidative stress than Pparb/dfl/fl tumors. Similarly, the colorectal cancer cells and human colon epithelial cells also experienced lower oxidative stress when co-cultured with fibroblasts depleted of PPARβ/δ expression. Therefore, our results establish a role for fibroblast PPARβ/δ in epithelial–mesenchymal communication for ROS homeostasis.
Similar content being viewed by others
Introduction
Tumor initiation, growth, and propagation are frequently accompanied by desmoplasia and acquisition of a reactive stroma, mainly comprising leukocytes, fibroblasts, and endothelial cells [1]. The dynamic and reciprocal relationship between epithelial and mesenchymal compartments of the tumor dictates almost every aspect of cancer progression, even governing the efficacy of therapy and influencing the risk of disease relapse [2]. This epithelial–mesenchymal communication has been usually ascribed to a complex signaling network of growth factors and cytokines. Reactive oxygen species (ROS) have also been identified as potent modifiers of signaling pathways and consequently cell behavior. The balance between production of ROS and their elimination by antioxidants is important for tissue homeostasis [3, 4]. Microenvironmental oxidative stress conjures protective antioxidant response from tissues to regain homeostasis. However, ROS imbalance or the loss of homeostatic control in a continued oxidative microenvironment can either result in cell death caused by oxidative catastrophe or contribute to carcinogenesis, which are tissue- and context-dependent [5,6,7]. Indeed, ROS are by-products of aberrant cancer cell metabolism and mitochondrial defects. However, little is known about the effect of tumor modulating ROS signals by fibroblasts, the most prominent cell type in tumor microenvironment.
Recent study showed that a deficiency in fibroblast peroxisome proliferator-activated receptor β/δ (PPARβ/δ) increased oxidative stress in wound microenvironment to delay keratinocyte migration [8]. PPARβ/δ has also been implicated in the pathogenesis of colorectal cancer (CRC). However, its role remains controversial as both pro- and anti-tumorigenic roles was reported from studies using conventional PPARβ/δ-knockout mice [9, 10]. On one hand, PPARβ/δ has been shown to exert potent pro-tumorigenic effects. Early study suggested that nonsteroidal anti-inflammatory drugs (NSAIDs) reduced tumorigenesis through inhibition of PPARβ/δ [11]. PPARβ/δ has been shown to promote UV-induced skin tumorigenesis by upregulating Src expression and activity thereby enhancing the downstream EGFR/Erk1/2 pathway [12]. PPARβ/δ also enhanced CRC progression by regulating Glut1 and SLC10A5 expression, promoting cancer cell metabolic programming [13]. PPARβ/δ also promoted angiogenesis by stimulating the expression of VEGF in CRC and bladder cancer [14,15,16]. In addition, PPARβ/δ promoted a pro-tumorigenic inflammatory environment through the increased production of COX-2-derived PGE2 [16]. On the other hand, PPARβ/δ has also been reported to exert potent anti-inflammatory functions through interfering with the functions of transcription factors such as NFκB, AP1, STAT3 [17]. In melanoma, PPARβ/δ exhibited inhibitory effects on tumorigenicity via the modulation of inflammatory and apoptotic mechanisms [18]. In apparent discrepancies with earlier studies, several studies showed that NSAIDs increased PPARβ/δ expression in human CRC cell lines, suggesting that elevated PPARβ/δ level, rather than decreased level, contributed to the chemopreventive effects of NSAIDs. Using a tissue-specific approach to delineate the problem, subsequent studies using an intestinal epithelia-specific deletion and transgene overexpression of PPARβ/δ in mice supported a pro-tumorigenic role for epithelial PPARβ/δ [19, 20]. Recent work on skin squamous cell carcinoma showed that the expression of PPARβ/δ was reduced in cancer-associated fibroblasts [21]. Further investigation revealed that PPARβ/δ in cancer-associated fibroblasts reduced the invasiveness, xenobiotic resistance and cellular bioenergetics of the adjacent epithelial cancer, at least, in part via the secretion of growth factors and cytokines [21]. However, the role of fibroblast PPARβ/δ in the initiation and progression of CRC remains unclear.
Our understanding on the role of fibroblasts in pathologic conditions is hampered by the absence of specific markers. Fibroblast-specific protein (FSP)1 (or S100A-4) has been suggested as a fibroblast-specific marker in normal and fibrotic tissues. The intestinal mesenchyme comprises of a heterogeneous population of stromal cells of varying origins [22]. FSP1-directed deletion occurs in fibroblast subpopulation that had an active FSP1 promoter activity. Although FSP1 has been identified as a fibroblast marker and used for fibroblast depletion [23, 24], the complete coverage of the entire fibroblast population by a single marker has proven to be tricky. Although FSP1 is only expressed in the mesenchymal lineage and commonly used for stromal-specific gene deletion strategy, it is lowly expressed if at all, in matured fibroblasts and myofibroblasts. Despite these limitations, FSP1 reporter mice and FSP1-Cre-driven gene deletion are considered reliable strategies to investigate fibroblast biology. Transgenic mice with green fluorescent protein expression driven by the FSP1 promoter have been used to map cell fate in fibroblast populations, and that FSP1-Cre animals have served as important tools to dissect the effects of fibroblast-specific gene deletion in fibrotic and neoplastic conditions. FSP1-directed deletion of genes, like Smoothened, PTEN, TGF receptor type II, and BMP receptor 2 in the fibroblasts have been used to study their roles in different tumor types such as acinar–ductal metaplasia, prostate, breast, and squamous cell carcinoma of the forestomach [25,26,27,28].
Drawing on the parallels between wound healing and cancer, we investigated the role of fibroblast PPARβ/δ in CRC. We generated a mouse whose PPARβ/δ gene was deleted in the fibroblasts (FSPCre-Pparb/d−/−) by crossing a floxed PPARβ/δ mouse (Pparb/d fl/fl) [29] with transgenic mice with a Cre transgene under the control of FSP1 promoter (FSP1-Cre) [30]. The FSPCre-Pparb/d−/− mice developed fewer CRC when compared to their Pparb/dfl/fl littermates, regardless of genetic-, or chemical-induced carcinogenesis. Microarray gene expression analyses revealed an altered oxidative processes in adjacent intestinal or tumor epithelium of FSPCre-Pparb/d−/− mice. We recapitulated the reduced oxidative stress phenotype in two human intestinal cancer cell lines (HCT116 and HT29) when co-cultured with colonic fibroblasts from FSPCre-Pparb/d−/− mice or fibroblasts whose endogenous PPARβ/δ were suppressed by siRNA. Taken together, our results indicate that PPARβ/δ expression in fibroblast modulates oxidative responses in the adjacent epithelium, and oxidative stress may be an initiating factor in CRC.
Results and discussion
PPARβ/δ expression is ablated in colonic fibroblasts of FSPCre-Pparb/d −/− mice
We generated a transgenic mouse line harboring a fibroblast-specific deletion of PPARβ/δ exon 4 allele (FSPCre-Pparb/d−/−) (Sng et al., in revision). To study fibroblast PPARβ/δ ablation in the gastrointestinal tract, we examined the deletion PPARβ/δ exon 4 in the different colonic tissue layers of the FSPCre-Pparb/d−/− transgenic mice by dissecting the mucosa villi, stroma laminal propria and muscularis layers with a laser dissecting microscope (Fig. 1a; Supplementary Fig. 1A–D). Subsequent PCR and DNA gel analysis confirmed the deletion of exon 4 of PPARβ/δ gene has occurred in the intestinal fibroblasts, which were enriched in the locality of the stroma lamina propria region (Fig. 1b). To examine PPARβ/δ protein expression in colonic stroma, we enriched for colonic fibroblasts from FSPCre-Pparb/d−/− and their Pparb/dfl/fl littermates by the fibroblast explant method. Our analysis of explanted FSPCre-Pparb/d−/− fibroblasts showed a 60 and 80% decrease in PPARβ/δ protein and mRNA levels, respectively, when compared with Pparb/dfl/fl littermates (Fig. 1c, d). The observed residual PPARβ/δ expression may be contributed by two technical difficulties. As mentioned above, FSP1 promoter is active in a subpopulation of fibroblasts. Although, the explant method is a reliable method to enrich and culture primary fibroblasts in vitro, it is also inevitable that other cell types such as endothelial cells and inflammatory immune cells from the stroma might have contaminated the culture [22, 31]. Importantly, our observation mirrored that in other studies where FSP1 promoter-driven gene deletion was targeted to the stroma of breast and pancreatic tissues [26, 30]. Taken together, we have established that the FSPCre-Pparb/d−/− colonic mesenchyme harbors perturbed PPARβ/δ expression.
PPARβ/δ expression is ablated in colonic fibroblasts of FSPCre-Pparb/d−/− mice. a Representative H&E-stained images of colons before and after laser capture microdissection of colonic layers, mucosal, lamina propria, and muscularis. FSPCre-Pparb/d−/− mice were generated in-house by crossing Pparb/dfl/fl and FspCre mice progenies, and backcrossed with Pparb/dfl/fl for at least six generations. Scale: 500 µm (b) Schematic diagram depicting the relative position of PCR genotyping primers for floxed and deleted Pparb/d exon 4 alleles, at 400 bp (red), and 490 bp (blue), respectively (top). Sample DNA were processed as instructed in the KAPA Express Extract kit and 2 × KAPA2G Fast Genotyping Mix. DNA gel image showing genotyping results indicated tissue layers colonic submucosa and lamina propria layer (L.p.); mucosal villi (Vil.); smooth muscle (Mus.) layers. c Representative images showing the outgrowth of fibroblasts from colonic explants from Pparb/dfl/fl and FSPCre-Pparb/d−/− mice. Fresh colons were dissected longitudinally and washed in ice cold phosphate buffered solution (PBS) containing 30% antimycotic/antibiotic solution. Specimens were minced to 1–2 mm2 pieces and incubated in fresh culture media for 72 h to allow colonic fibroblasts to migrate out of the explants. Migrated cells were washed and cultured for another 48 h before protein and RNA analyses. Scale bar: 500 µm. c–d Relative fold change in expression of PPARβ/δ mRNA c and PPARβ/δ protein d in migrated colonic fibroblasts. Total RNA was extracted from cells using TRIzol Reagent with E.Z.N.A Total RNA Kit according to manufacturer’s protocol. Reverse transcription real-time quantitative PCR (RT-qPCR) were performed with 18S rRNA as a housekeeping gene. Samples were homogenized and lysed in mammalian protein extraction reagent (M-PER) supplemented with complete protease inhibitor mix. Far-infrared immunoblotting was performed. β-tubulin that served as housekeeping protein was from the same samples. Data are represented as mean ± S.E.M. from at least four independent experiments
Loss in fibroblast PPARβ/δ expression delays intestinal tumorigenesis
To investigate the effects of fibroblast PPARβ/δ expression on intestinal tumorigenesis, we employed three commonly used murine models, chemical (azoxymethane, AOM, and dextran sulfate sodium, DSS), genetic (APCmin/+), and combined (APCmin/+ with DSS) approaches to induce colon tumors in FSPCre-Pparb/d−/− mice.
Ulcerative colitis and inflammatory bowel syndrome are common risk factors associated with CRC development [32, 33]. Well-established animal models for ulcerative colitis and inflammatory bowel syndrome have used DSS to induce colitis in mice, thus DSS was used to induce intestinal inflammation in two of the murine models. We investigated whether there were differences in the inflammatory processes that lead to tumor initiation. In DSS-induced colitis, the FSPCre-Pparb/d−/− mice yielded a lower disease activity index (DAI) (Fig. 2a and Supplementary Table 2), reduced DSS-induced morphological aberrations and loss of the mucosa layer (Fig. 2b, c) when compared with wild-type littermates. No significant difference in colon lengths was observed (Fig. 2d). Consistent with the lower disease severity, we detected a reduction of pro-inflammatory cytokines and an increase of anti-inflammatory cytokines in FSPCre-Pparb/d −/− when compared with Pparb/d fl/fl mice as determined by multiplex immunoassay (Supplementary Table 1). These observations suggest that a deficiency in fibroblast PPARβ/δ confers some protection against colitis, possibly by modulating an inflammatory response.
Loss in fibroblast PPARβ/δ expression retards intestinal tumorigenesis. a Mean disease activity index (DAI) score of DSS-treated mice over 7 days. Litter-matched mice were fed with 2% DSS in drinking water ad libitum and observed at days 3, 5, and 7. DAI score was computed as detailed in Supplementary Materials and Methods. Values are mean ± S.E.M. (n = 5 for each genotype per condition). b Representative H&E stained images of mouse colons and ileums from Pparb/dfl/fl and FSPCre-Pparb/d−/− in vehicle- and DSS-treated conditions. Scale: 100 µm. c Mean villus number measurement of mouse ilea in Vehicle and DSS-treated mice. H&E stained images of mouse ilea (n = 8) were used for the recording of villus number in ImageJ software version 1.45 s. d Length of colons from Pparb/dfl/fl and FSPCre-Pparb/d−/− mice after treatment with vehicle or DSS. Each data point represents one mouse. e Percentage survival of mice after AOM/DSS treatment for 10 weeks. Mice were injected with 10 mg/kg AOM. After 1 week, they were given 2% DSS in drinking water ad libitum. Litter-matched FSPCre-Pparb/d−/− mice (n = 13) and Pparb/dfl/fl mice (n = 18) were used. f Mean DAI score of AOM/DSS-treated FSPCre-Pparb/d−/− mice (n = 13) and Pparb/dfl/fl littermates (n = 18). DAI score was computed as detailed in Supplementary Materials and Methods. Values are mean ± S.E.M. g Representative images of colons from vehicle- and AOM/DSS-treated Pparb/dfl/fl and FSPCre-Pparb/d−/− mice. Colons were dissected longitudinally from the distal to proximal end (right to left) and inspected for tumor number and size using an upright dissecting microscope. Scale: 1 cm. h Bar graph depicting total colonic tumor numbers per mouse in AOM/DSS-treated FSPCre-Pparb/d−/− mouse colons (n = 6) and cognate Pparb/dfl/fl colons (n = 6). Values are mean ± S.E.M. i Box-and-whisker plot showing distribution of tumor number and size in AOM/DSS-treated Pparb/dfl/fl and FSPCre-Pparb/d−/− mice. The tumors were categorized into three groups by tumor diameter (small < 2 mm; middle, 2–4 mm, large > 4 mm). Fewer large tumors were observed in FSPCre-Pparb/d−/− colons than Pparb/dfl/fl colons. Values are mean ± S.E.M. (n = 6 for each genetic background). j Representative H&E-stained images of tumors and adjacent colon tissues from FSPCre-Pparb/d−/− and Pparb/dfl/fl mice. Scale: 200 µm
Next, we looked at chemical induction of colon tumors using AOM/DSS treatment. In similar fashion to our colitis experiment, the FSPCre-Pparb/d−/− mice demonstrated better survival and DAI over the course of AOM/DSS treatment, compared to the wild-type (Pparb/dfl/fl) mice (Fig. 2e, f). The FSPCre-Pparb/d−/− mice also produced 50% fewer and smaller tumors when compared with Pparb/dfl/fl littermates (Fig. 2g–j). The results showed that the FSPCre-Pparb/d−/− genetic background confers a protection against CRC in mice, suggesting that fibroblast PPARβ/δ is important in the promotion of tumor development.
To study intestinal tumor formation in a spontaneous CRC murine model, we crossed FSPCre-Pparb/d−/− mice to an APCmin/+ background (Supplementary Figure 1E–M). APCmin/+ mice were either allowed to spontaneously develop tumors or stimulated chemically by a 1-week DSS treatment course. First, we noted a lower DAI in APCmin/+ transgenic mice on the FSPCre-Pparb/d−/− background (Supplementary Figure 1E & F). In both models, we also noted that mice deficient in fibroblast PPARβ/δ were more resistant to the disease burden of tumor development (Supplementary Figure 1G) as suggested by higher survival rates (Supplementary Figure 1H & K). The mice deficient in fibroblast PPARβ/δ also developed fewer and smaller tumors compared to cognate wild-type Pparb/dfl/fl littermates (Supplementary Figure 1G, I–J & L-M). Thus, regardless of the oncogenic stimuli, either with AOM/DSS, APCmin or combined, our results indicate that reduced PPARβ/δ expression in fibroblasts delays CRC development.
Fibroblast PPARβ/δ alters NRF2-mediated response in FSPCre-Pparb/d −/− intestinal epithelium
To gain mechanistic insight into how PPARβ/δ expression in fibroblasts mediates changes in its adjacent epithelial cells, we performed a comparative gene expression analysis of epithelial and mucosal layers from normal intestines and tumors from FSPCre-Pparb/d−/− and APCmin/+FSPCre-Pparb/d−/−. Ingenuity pathway analyses revealed that genes involved in oxidative phosphorylation and mitochondrial dysfunction were enriched in FSPCre-Pparb/d−/− mice (Fig. 3a–c). Interestingly, our upstream regulator analyses of normal tumor adjacent and tumor epithelia revealed enhanced NRF2-mediated stress responses owing to the deletion of fibroblast PPARβ/δ (Fig. 3d, e). NRF2 is a master regulator of antioxidant responses through binding to the antioxidant response elements in the gene’s promoter region. It upregulates the expression of numerous antioxidant proteins to protect the cell from oxidative damages [34]. Available experimental evidence also clearly indicates that NRF2 is an important player in the maintenance of mitochondrial homeostasis and structural integrity (Holmstrom, Kostov et al. 2016). This role becomes particularly critical under conditions of oxidative, electrophilic, and inflammatory stress which influences the overall health and survival of the organism. The loss of NRF2 in mouse has been associated with increased susceptibility to CRC development [35], underscoring NRF2 as a potential chemopreventive target [36]. PPARs have been implicated as one of the downstream mediators of NRF2 [37]. For example, functional PPAR response element has been identified in the glutathione S-transferase [38, 39], glutathione peroxidase 1, and catalase genes [8]. PPARβ/δ has also been shown to exert antioxidant effect on cardiomyocytes by inhibiting PI3K/Akt signaling pathway to suppress ROS generation induced by AngII, however very little is known about its role in redox regulation in the gut [40,41,42]. Therefore, we winnowed our attention on NRF2 and associated antioxidant genes in the colon of FSPCre-Pparb/d−/− mice.
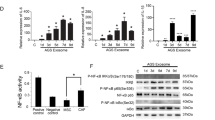
Fibroblast PPARβ/δ alters NRF2 and associated oxidative processes in FSPCre-Pparb/d−/− intestinal epithelium. a–c Gene Ontological Pathway Analysis of differentially expressed genes from colon mucosa (a), ileum mucosa (b), and intestinal tumor epithelium (c). Total RNA was isolated from LCM tissues using the Ambion RecoverAll Total Nucleic Acid Isolation Kit for FFPE and processed for array hybridization using the GeneChip WT Pico Kit and GeneChip Mouse Gene 1.0 ST Array gene chips. The data were normalized and analyzed using the Partek Genomics Suite v6.6. Multi-way ANOVA analysis was used to compare FSPCre-Pparb/d−/− (KO) against Pparb/dfl/fl (WT). Gene ontology (GO) analysis of gene subsets was performed using the Ingenuity Pathway Analysis software. The top five GO analysis were in descending order as determined by their p-value. Pathway analyses were conducted using Ingenuity Pathway Analysis (IPA), comparing FSPCre-Pparb/d−/− (KO) against Pparb/dfl/fl (WT) samples (n = 3; Fold change cutoff ≥ ± 1.3; p < 0.05). * Oxidative Phosphorylation and Mitochondrial Dysfunction were two common pathways placed in the top three most significant pathways across all tissues. d, e Unguided hierarchical clustering by upstream regulator (d) and pathway analysis (e) of differentially expressed genes in normal, tumor adjacent and tumor epithelium tissues from FSPCre-Pparb/d−/− (KO) and Pparb/dfl/fl (WT) mice. Our analysis also flagged NRF2-mediated oxidative stress response as an upregulated pathway in FSPCre-Pparb/d−/− colonic epithelium. *NFE2L2 (Nuclear factor erythroid 2-related factor 2) is a synonym of NRF2 (Nuclear factor erythroid 2-related factor 2). f Intracellular H2O2 levels in colons from FSPCre-Pparb/d−/− and Pparb/dfl/fl mice as determined by Amplex Red H2O2 assay. Samples were homogenized and supernatants incubated with appropriate reagents according to manufacturer’s instructions. Oxidation of Amplex Red results in the production of fluorescent resorunfin (571 nm ex and 585 nm em). A calibration curve was prepared (0–20 μM) and used to calculate H2O2 concentration of samples. The concentration of H2O2 was normalized to total protein. Data are represented as mean ± S.E.M. from at least four independent experiments. g Relative fold change in mRNA levels of 13 genes associated with oxidative stress response in APCmin/+FSPCre-Pparb/d−/− and APCmin/+Pparb/dfl/fl colons. Total RNA was extracted, and RT-qPCR was performed as described in legends of Fig. 1d, e. Ribosomal RNA L27 used as housekeeping gene. Values are mean ± S.E.M. (n = 4). *p < 0.05. h Representative images of immunohistochemical staining for NRF2 (black arrows) in FSPCre-Pparb/d−/− and Pparb/dfl/fl colons. Sections were counterstained with hematoxylin for visualization of tissue morphology. The number of NRF2-positive cells was recorded using ImageJ software version 1.45 s. Black arrows indicate representative NR2-positive regions. Scale bar: 50 µm. i Fold change in NRF2 protein in colons from FSPCre-Pparb/d−/− and Pparb/dfl/fl mice. Representative immunoblot and graph for NRF2 protein are shown. Samples were processs for western bllot as described in legends of Fig. 1d, e. PCNA that served as housekeeping protein were from the same samples. Data are represented as mean ± S.E.M. from at least three independent experiments
We observed an increase in NRF2 expression, concomitant with elevated expression of antioxidant enzymes in intact FSPCre-Pparb/d−/− colons (Fig. 3f). We also found higher expression of antioxidant (e.g., glutathione peroxidase and catalase) genes in the FSPCre-Pparb/d−/− compared with Pparb/dfl/fl intestines (Fig. 3g). This was supported by increased NRF2 staining in the intestinal villi, as well as increased nuclear NRF2 protein in FSPCre-Pparb/d−/− colons (Fig. 3h–i). Indeed, Therefore, our data provides evidence that NRF2 and its downstream oxidative stress signaling pathways were elevated in FSPCre-Pparb/d−/− colons and may be a major contributing factor for the reduced CRC in these mutant mice.
PPARβ/δ deficiency in fibroblasts lowers oxidative stress in HCT116, HT29, and iCEC cells
As shown earlier in the inflammatory model, FSPCre-Pparb/d−/− colons had a less-inflamed phenotype, with more anti-inflammatory cytokines and less-inflammatory cytokines, compared with their Pparβ/δfl/fl littermates. Inflammatory immune cells release a myriad cytokines, growth factors, and ROS to create an inflammatory microenvironment that is conducive for tumor development [43, 44]. Thus, we questioned if infiltrating immune cells could contribute to the difference in the tumor load between FSPCre-Pparb/d−/− and Pparb/dfl/fl mice. We did not observe differences in the number of CD11b+ immune cell numbers between two genotypes (Fig. 4a, b). Although the total number of infiltrating CD11b+ immune cells was similar, multiplex immunoassay analysis (Supplementary Table 1) suggested potential differences in the relative abundance of neutrophils, T-cells and macrophages between FSPCre-Pparb/d−/− and Pparb/dfl/fl colons. We did not find any significant difference in the number of infiltrating neutrophils (Ly6G+CD11b+) and T lymphocytes (CD3+CD11b+) between the FSPCre-Pparb/d−/− and Pparb/dfl/fl colons as determined by FACS analysis (Supplementary Fig. 2A & B). We noted that the vehicle-treated FSPCre-Pparb/d−/− colons have a higher infiltrated macrophage (F4/80+Cd11b+) than Pparb/dfl/fl, which was no longer distinguishable upon DSS stimulation (Supplementary Fig. 2C). Further characterization of macrophage subtypes revealed that vehicle-treated FSPCre-Pparb/d−/− colons have more of the anti-inflammatory M2 macrophages, but higher M1 macrophages upon DSS treatment (Supplementary Fig. 2D). The lower M1:M2 ratio in FSPCre-Pparb/d−/− colon was consistent with the reduced inflammatory cytokine landscape. Clinical and experimental evidence have indicated that inflammatory milieu and elevated ROS contribute to cancer initiation [45, 46]. ROS production is usually associated with the activation and functions of M1 rather than M2 macrophages. Together, they suggest that immune cells have minor role in tumor initiation of FSPCre-Pparb/d−/− mice.
PPARβ/δ deficiency in fibroblasts lowers oxidative stress in colon cell lines. a, b FACS analysis of CD11b + cell numbers in colons from FSPCre-Pparb/d−/− and Pparb/dfl/fl mice treated with vehicle or DSS (a) and APCmin/+FSPCre-Pparb/d−/− and APCmin/+Pparb/dfl/fl colons (b). Fresh tissue biopsies were dissociated using the gentleMACS Dissociator according to manufacturer’s instruction. Homogenates were strained, washed, and processed for staining with fluorophore-conjugated antibodies on ice. Flow cytometry was performed using Accuri C6 Flow Cytometer and analyzed on FlowJo v10.0.7. Values are mean ± S.E.M. (n = 6). c Extracellular H2O2 levels of explanted fibroblasts from Pparb/dfl/fl and FSPCre-Pparb/d−/− colons detected by Amplex Red Assay as described in legend of Fig. 3f. Values are mean ± S.E.M. (n = 6). d, e Intracellular H2O2 levels of explanted fibroblasts from Pparb/dfl/fl and FSPCre-Pparb/d−/− colons detected by CellROX. Cells were first positively gated for fibroblast marker (CD140a or PDGRA) and the CellROX fluroscence readings were taken from the CD140a + gated cells. Values are mean ± S.E.M. (n = 4). fRelative fold change in mRNA levels of 13 genes associated with oxidative stress response in explanted fibroblasts from Pparb/dfl/fl and FSPCre-Pparb/d−/− colons. Total RNA was extracted, and RT-qPCR was performed as described in legends of Fig. 1d, e. were Ribosomal 18S rRNA served as a housekeeping gene. Data are represented as mean ± S.E.M. from 4 independent experiments. * p < 0.05, ** p < 0.01. g, h, k, n Relative fold change in mRNA levels of 13 genes associated with oxidative stress response in HCT116 (g, k, n) and HT29 cells (h) cultured in conditioned medium of fibroblasts from Pparb/dfl/fl and FSPCre-Pparb/d−/− (g, h), or co-cultured with BJ-1 (k) and CCD18Co cells (n) whose endogenous PPARβ/δ was suppressed by siRNA. Cells transfected with siRNA of PPARβ/δ and Scrambled were denoted by superscript. Data are represented as mean ± S.E.M. from four independent experiments. * p < 0.05, ** p < 0.01. i, l, o Intracellular oxidative stress levels in HCT116 cells (i, l, o) either cultured in condition medium of fibroblasts from Pparb/dfl/fl and FSPCre-Pparb/d−/− (i) or co-cultured with BJ-1 (l) and CCD18Co cells (o) whose endogenous PPARβ/δ was suppressed by siRNA. Cells transfected with Scrambled siRNA served as cognate controls. Intracellular ROS level of HCT116 cells cultured in conditioned medium from or co-cultured with PPARβ/δ-deficient fibroblasts as determined by CM-H2DCFDA assay. Values are mean ± S.E.M. (n = 4). j, m Relative fold change in PPARβ/δ mRNA expression in BJ-1 fibroblasts (j) and CCD18co myofibroblasts (m) whose endogenous PPARβ/δ was suppressed by siRNA at 48 and 72 h post transfection. Cells transfected with Scrambled siRNA served are cognate controls. 18 S rRNA served as housekeeping gene. Data are represented as mean ± S.E.M. from four independent experiments. * P < 0.05, ** P < 0.01
Previous study reported that PPARβ/δ in wound fibroblasts transcriptionally regulates the expression of H2O2-detoxifying enzymes, glutathione peroxidase 1 and catalase, culminating in reduced ROS in the wound microenvironment [8]. Indeed, we detected elevated extracellular H2O2 in the conditioned medium of PPARβ/δ-deficient fibroblasts compared to their wild-type counterparts (Fig. 4c). No significant difference in intracellular H2O2 levels in the fibroblasts (Fig. 4d, e). Explanted FSPCre-Pparb/d−/− fibroblasts also have overall reduction in the expression antioxidant genes (8 of 12 genes), suggesting a more oxidative phenotype that contributed to the elevated H2O2 production and secretion into the microenvironment (Fig. 4f). Numerous studies have showed that the activation of cytoprotective NRF2-mediated stress responsive genes in response to elevated oxidative stress, like extracellular H2O2. Thus, to strengthen our observation that the novel epithelial–mesenchymal communication in oxidative stress homeostasis was dysregulated in FSPCre-Pparb/d−/−, as well as a direct effect of fibroblast PPARβ/δ on the oxidative status of adjacent epithelia, we measured ROS in two different human intestinal cancer cell lines (HCT116 and HT29) co-cultured with either PPARβ/δ-deficient fibroblasts or in conditioned media from FSPCre-Pparb/d−/− and Pparb/d−/− colonic fibroblasts (Fig. 4g–o). Consistent with our gene expression data, we detected higher expression of NRF2 and associated antioxidant enzymes in HCT116 and HT29 cultured in conditioned media from FSPCre-Pparb/d−/− than in PPARb/dfl/fl colonic fibroblasts (Fig. 4g, h). This is coupled with reduced ROS levels in HCT116 cells cultured in conditioned media from FSPCre-Pparb/d−/− colonic fibroblasts (Fig. 4i). In support, we also detected higher NRF2 expression, elevated mRNA levels of antioxidant enzymes and reduced ROS levels in HCT116 cells when co-cultured with siPPARβ/δ-knockdown BJ-1 fibroblasts (BJ-1siPPARβ/δ) (Fig. 4 j–l) or CCD18Co myofibroblasts (CCD18CosiPPARβ/δ) (Fig. 4m–o)compared with cognate controls (BJ-1siScrambled and CCDCosiScrambled). We similarly observed an NRF2 antioxidant response in immortalized non-tumorigenic human colon epithelial cells cultured in either conditioned media from FSPCre-Pparb/d−/− or the presence of exogenous H2O2 (Supplementary Figure 2 E-G). This response was attenuated in the presence of antioxidant N-acetyl cysteine (NAC). Chronic or repeated exposure to low level of H2O2, in combination with growth factors, has been shown to exert transformative and growth-promoting effects on epithelial cells [47]. We observed an increase in HCT116 cells at either G2/M or S phase when was co-cultured with PPARβ/δ-knockdown fibroblast cell lines compared with cognate controls (Supplementary Figure 2H).
Our findings suggest that an elevated NRF2-related response in the colon epithelia was associated with reduced CRC. Thus, we explore the effect of NAC pre-treatment on CRC initiation in DSS-treated APCmin/+Pparb/dfl/fl and APCmin/+FSPCre-Pparb/d−/− mice (NAC/DSS treatment). APCmin/+Pparb/dfl/fl mice developed more and larger CRCs than APCmin/+FSPCre-Pparb/d−/− (Supplementary Figure 1L-M). The pre-treatment with NAC reduced the number of tumors in APCmin/+Pparb/dfl/fl by 87% when compared no NAC pre-treatment (Supplementary Figure 1N-P), suggesting that ROS have an important role in early tumor development. There was no difference in the number of tumors between NAC pre-treated or untreated APCmin/+FSPCre-Pparb/d−/− mice (Supplementary 1L & O), suggesting that further reduction in epithelial ROS has minimal impact on tumor development. Paradoxically, we observed a reduced median survival rate of the APCmin/+FSPCre-Pparb/d−/− mice pre-treated with NAC (Fig. 1n). Although the precise reason for this observation is unclear, we speculate that additional reduction in epithelial ROS might have disrupted critical ROS-mediated cellular functions necessary for homeostasis in APCmin/+FSPCre-Pparb/d−/− mice, which already have elevated NRF2 antioxidant response. Studies from the past two decades have established that ROS serve as signaling molecules to regulate biological and physiological processes [48]. Clearly, future experiments will be necessary to understand the effect of antioxidant treatment on normal cellular functions in FSPCre-Pparb/d−/− mice, as well as the optimal antioxidant treatment regime to reduce CRC in wild-type mice.
We showed a major role for stromal PPARβ/δ in ROS production that consequently conjures an NRF2-mediated antioxidant response, reducing ROS levels in the adjacent intestinal epithelial cancer cells. NRF2-mediated response also results in the activation of various signaling pathways that facilitates tumor cell senescence [49, 50]. Collectively, we propose that FSPCre-Pparb/d−/− colonic epithelium are subjected to a constant dose of ROS, extracellular H2O2 from their neighboring stroma, which triggers NRF2-mediated signaling that suppresses tumor development.
Conclusion
Our data have shown that PPARβ/δ ablation in fibroblasts results in reduced tumorigenesis across three different mice models of CRC. Microarray gene expression and gene ontology analyses, together with in vivo and in vitro experiments indicate that PPARβ/δ depletion in fibroblasts alters the antioxidant responses and thus the oxidative status of the adjacent epithelium. We propose that PPARβ/δ deficiency in fibroblasts increases extracellular H2O2, triggering an NRF2-mediated antioxidant response in the adjacent epithelia. The elevated expression of NRF2-dependent proteins is critical for eliminating carcinogens to maintain cellular redox homeostasis. Consequently, FSPCre-Pparb/d−/− mice have reduced colonic polyp formation.
Nuclear receptor PPARβ/δ has been implicated in CRC, although it remained controversial as studies have shown supporting evidence for PPARβ/δ playing an anti-tumorigenic [16] and pro-tumorigenic roles in CRC [51]. It is conceivable that PPARβ/δ has dual roles in tumorigenesis, much like TGF-β1 and ROS. By regulating cell growth, death, and immortalization, TGFβ signaling pathways exert tumor suppressor effects in normal cells and early carcinomas. But as tumors progress, these protective and cytostatic effects of TGFβ are often lost, switching to promote cancer progression, invasion, and tumor metastasis. Similarly, chronic oxidative stress has been shown to promote tumorigenesis [47, 52], whereas the modulation of oxidative stress as an anticancer therapeutic agent has also been discussed [53]. With the temporal and dose-dependent basis of oxidative stress on tumor formation and development, this may explain the dual effect of PPARβ/δ on tumorigenesis.
Limitations of our study include that one genetic background of mouse and our deletion strategy consists of the deletion of exons coding for the DNA-binding domain of PPARβ/δ. Different mouse strains may exhibit different susceptibility to carcinogen or tolerance to oxidative stress. It is also conceivable that other gene deletion strategies may result in different phenotypic severity or outcomes owing to differences in vulnerabilities to the oxidative stress. FSP1 is a key marker of a specific subset of macrophages in the liver during fibrosis and injury [54], although no report has described confounding issues in other organs. Nevertheless, to the best of our knowledge, this is the first study to examine the role of fibroblast PPARβ/δ in CRC, which reveals novel epithelial–mesenchymal communication in ROS homeostasis. Our results illustrate that PPARβ/δ modulation of oxidative stress has potential medical applicability.
References
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37.
Udagawa T, Wood M. Tumor-stromal cell interactions and opportunities for therapeutic intervention. Curr Opin Pharmacol. 2010;10:369–74.
Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21.
Manda G, Isvoranu G, Comanescu MV, Manea A, Debelec Butuner B, Korkmaz KS. The redox biology network in cancer pathophysiology and therapeutics. Redox Biol. 2015;5:347–57.
Chaiswing L, Oberley TD. Extracellular/microenvironmental redox state. Antioxid Redox Signal. 2010;13:449–65.
Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sanchez-Perez P, Cadenas S, et al. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–97.
Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253.
Wang X, Sng MK, Foo S, Chong HC, Lee WL, Tang MB, et al. Early controlled release of peroxisome proliferator-activated receptor beta/delta agonist GW501516 improves diabetic wound healing through redox modulation of wound microenvironment. J Control Release. 2015;197:138–47.
Peters JM, Gonzalez FJ, Muller R. Establishing the role of PPARbeta/delta in carcinogenesis. Trends Endocrinol Metab. 2015;26:595–607.
Tan NS, Vazquez-Carrera M, Montagner A, Sng MK, Guillou H, Wahli W. Transcriptional control of physiological and pathological processes by the nuclear receptor PPARbeta/delta. Prog Lipid Res. 2016;64:98–122.
He TC, Chan TA, Vogelstein B, Kinzler KW. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–45.
Montagner A, Delgado MB, Tallichet-Blanc C, Chan JS, Sng MK, Mottaz H, et al. Src is activated by the nuclear receptor peroxisome proliferator-activated receptor beta/delta in ultraviolet radiation-induced skin cancer. EMBO Mol Med. 2014;6:80–98.
Zhang W, Xu Y, Xu Q, Shi H, Shi J, Hou Y. PPARdelta promotes tumor progression via activation of Glut1 and SLC1-A5 transcription. Carcinogenesis. 2017;38:748–55.
Marin HE, Peraza MA, Billin AN, Willson TM, Ward JM, Kennett MJ, et al. Ligand activation of peroxisome proliferator-activated receptor beta inhibits colon carcinogenesis. Cancer Res. 2006;66:4394–401.
Wang D, Wang H, Guo Y, Ning W, Katkuri S, Wahli W, et al. Crosstalk between peroxisome proliferator-activated receptor delta and VEGF stimulates cancer progression. Proc Natl Acad Sci USA. 2006;103:19069–74.
Wang D, Fu L, Ning W, Guo L, Sun X, Dey SK, et al. Peroxisome proliferator-activated receptor delta promotes colonic inflammation and tumor growth. Proc Natl Acad Sci USA. 2014;111:7084–9.
Kilgore KS, Billin AN. PPARbeta/delta ligands as modulators of the inflammatory response. Curr Opin Investig Drugs. 2008;9:463–9.
Borland MG, Yao PL, Kehres EM, Lee C, Pritzlaff AM, Ola E, et al. Editor’s highlight: PPARbeta/delta and PPARgamma inhibit melanoma tumorigenicity by modulating inflammation and apoptosis. Toxicol Sci. 2017;159:436–48.
Zuo X, Peng Z, Moussalli MJ, Morris JS, Broaddus RR, Fischer SM, et al. Targeted genetic disruption of peroxisome proliferator-activated receptor-delta and colonic tumorigenesis. J Natl Cancer Inst. 2009;101:762–7.
Zuo X, Xu M, Yu J, Wu Y, Moussalli MJ, Manyam GC, et al. Potentiation of colon cancer susceptibility in mice by colonic epithelial PPAR-delta/beta overexpression. J Natl Cancer Inst. 2014;106:dju052.
Chan JSK, Sng MK, Teo ZQ, Chong HC, Twang JS, Tan NS. Targeting nuclear receptors in cancer-associated fibroblasts as concurrent therapy to inhibit development of chemoresistant tumors. Oncogene 2017. https://doi.org/10.1038/onc.2017.319
Pinchuk IV, Mifflin RC, Saada JI, Powell DW Intestinal mesenchymal cells. Curr Gastroenterol Rep. 2010;12:310–8.
Okada H, Inoue T, Kanno Y, Kobayashi T, Watanabe Y, Ban S, et al. Selective depletion of fibroblasts preserves morphology and the functional integrity of peritoneum in transgenic mice with peritoneal fibrosing syndrome. Kidney Int. 2003;64:1722–32.
Strutz F, Okada H, Lo CW, Danoff T, Carone RL, Tomaszewski JE, et al. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol. 1995;130:393–405.
Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–51.
Liu X, Pitarresi JR, Cuitino MC, Kladney RD, Woelke SA, Sizemore GM, et al. Genetic ablation of Smoothened in pancreatic fibroblasts increases acinar-ductal metaplasia. Genes Dev. 2016;30:1943–55.
Pickup MW, Hover LD, Polikowsky ER, Chytil A, Gorska AE, Novitskiy SV, et al. BMPR2 loss in fibroblasts promotes mammary carcinoma metastasis via increased inflammation. Mol Oncol. 2015;9:179–91.
Yu S, Yeh CR, Niu Y, Chang HC, Tsai YC, Moses HL, et al. Altered prostate epithelial development in mice lacking the androgen receptor in stromal fibroblasts. Prostate. 2012;72:437–49.
Iglesias J, Barg S, Vallois D, Lahiri S, Roger C, Yessoufou A, et al. PPARbeta/delta affects pancreatic beta cell mass and insulin secretion in mice. J Clin Invest. 2012;122:4105–17.
Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45.
Roncoroni L, Elli L, Doneda L, Piodi L, Ciulla MM, Paliotti R, et al. Isolation and culture of fibroblasts from endoscopic duodenal biopsies of celiac patients. J Transl Med. 2009;7:40.
Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639–45.
Kim ER, Chang DK. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol. 2014;20:9872–81.
Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–71.
Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila). 2008;1:187–91.
Saw CL, Kong AN. Nuclear factor-erythroid 2-related factor 2 as a chemopreventive target in colorectal cancer. Expert Opin Ther Targets. 2011;15:281–95.
Reddy RC, Standiford TJNrf2. and PPAR{gamma}: PPARtnering against oxidant-induced lung injury. Am J Respir Crit Care Med. 2010;182:134–5.
Cho HY, Gladwell W, Wang X, Chorley B, Bell D, Reddy SP, et al. Nrf2-regulated PPAR{gamma} expression is critical to protection against acute lung injury in mice. Am J Respir Crit Care Med. 2010;182:170–82.
Park EY, Cho IJ, Kim SG. Transactivation of the PPAR-responsive enhancer module in chemopreventive glutathione S-transferase gene by the peroxisome proliferator-activated receptor-gamma and retinoid X receptor heterodimer. Cancer Res. 2004;64:3701–13.
Kim HJ, Ham SA, Kim MY, Hwang JS, Lee H, Kang ES, et al. PPARdelta coordinates angiotensin II-induced senescence in vascular smooth muscle cells through PTEN-mediated inhibition of superoxide generation. J Biol Chem. 2011;286:44585–93.
Kim MY, Kang ES, Ham SA, Hwang JS, Yoo TS, Lee H, et al. The PPARdelta-mediated inhibition of angiotensin II-induced premature senescence in human endothelial cells is SIRT1-dependent. Biochem Pharmacol. 2012;84:1627–34.
Liu J, Wang P, He L, Li Y, Luo J, Cheng L, et al. Cardiomyocyte-restricted deletion of PPARbeta/delta in PPARalpha-null mice causes impaired mitochondrial biogenesis and defense, but no further depression of myocardial fatty acid oxidation. PPAR Res. 2011;2011:372854.
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–71.
Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22.
Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–96.
Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125:3347–55.
Chan JS, Tan MJ, Sng MK, Teo Z, Phua T, Choo CC, et al. Cancer-associated fibroblasts enact field cancerization by promoting extratumoral oxidative stress. Cell Death Dis. 2017;8:e2562.
Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Ramsey MR, Sharpless NE. ROS as a tumour suppressor? Nat Cell Biol. 2006;8:1213–5.
Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-delta attenuates colon carcinogenesis. Nat Med. 2004;10:481–3.
Khansari N, Shakiba Y, Mahmoudi M. Chronic inflammation and oxidative stress as a major cause of age-related diseases and cancer. Recent Pat Inflamm Allergy Drug Discov. 2009;3:73–80.
Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–47.
Osterreicher CH, Penz-Osterreicher M, Grivennikov SI, Guma M, Koltsova EK, Datz C, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc Natl Acad Sci USA. 2011;108:308–13.
Acknowledgements
EHPT and JSKC are recipients of the Nanyang President Graduate Scholarship; MKS is a recipient of the Research Scholarship of the NTU, Singapore. We wish to thank Pierre Chambon and Daniel Metzger for their help in generating the floxed PPARβ/δ mice. The authors also acknowledge Dr. Han Chung Chong and Ming Jie Tan of Denova Sciences Pte Ltd for assistance in microarray experiments and analysis.
Funding
This work was supported by Singapore Ministry of Education under Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2014-T2-1-012) and Academic Research Fund Tier 1 (RG134/15) to NST, and by the Lee Kong Chian School of Medicine, Nanyang Technological University Start-up Grant to WW.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/.
About this article
Cite this article
Tan, E.H.P., Sng, M.K., How, I.S.B. et al. ROS release by PPARβ/δ-null fibroblasts reduces tumor load through epithelial antioxidant response. Oncogene 37, 2067–2078 (2018). https://doi.org/10.1038/s41388-017-0109-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0109-8
This article is cited by
-
Deficiency in fibroblast PPARβ/δ reduces nonmelanoma skin cancers in mice
Cell Death & Differentiation (2020)
-
Exploiting vulnerabilities of cancer by targeting nuclear receptors of stromal cells in tumor microenvironment
Molecular Cancer (2019)
-
Adipose stromal cell targeting suppresses prostate cancer epithelial-mesenchymal transition and chemoresistance
Oncogene (2019)