Abstract
Slit-Robo GTPase-activating protein 1 (SRGAP1) functions as a GAP for Rho-family GTPases and downstream of Slit-Robo signaling. We aim to investigate the biological function of SRGAP1 and reveal its regulation by deregulated microRNAs (miRNAs) in gastric cancer (GC). mRNA and protein expression of SRGAP1 were examined by quantitative reverse transcription PCR (qRT-PCR) and western blot. The biological role of SRGAP1 was demonstrated through siRNA-mediated knockdown experiments. The regulation of SRGAP1 by miR-340 and miR-124 was confirmed by western blot, dual luciferase activity assays and rescue experiments. SRGAP1 is overexpressed in 9 out of 12 (75.0%) GC cell lines. In primary GC samples from TCGA cohort, SRGAP1 shows gene amplification in 5/258 (1.9%) of cases and its mRNA expression demonstrates a positive correlation with copy number gain. Knockdown of SRGAP1 in GC cells suppressed cell proliferation, reduced colony formation, and significantly inhibited cell invasion and migration. Luciferase reporter assays revealed that SRGAP1 knockdown significantly inhibited Wnt/β-catenin pathway. In addition, SRGAP1 was found to be a direct target of two tumor-suppressive miRNAs, miR-340 and miR-124. Concordantly, these two miRNAs were downregulated in primary gastric tumors and these decreasing levels w5ere associated with poor outcomes. Expression of miR-340 and SRGAP1 displayed a reverse relationship in primary samples and re-expressed SRGAP1, rescued the anti-cancer effects of miR-340. Taken together, these data strongly suggest that, apart from gene amplification and mutation, the activation of SRGAP1 in GC is partly due to the downregulation of tumor-suppressive miRNAs, miR-340 and miR-124. Thus SRGAP1 is overexpressed in gastric carcinogenesis and plays an oncogenic role through activating Wnt/β-catenin pathway.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a common malignancy worldwide, especially in East Asia countries. Most of the GC cases are adenocarcinomas and they are histologically further sub-divided into intestinal and diffuse subtypes. According to The Cancer Genome Atlas (TCGA), GC can be grouped into four molecular subtypes according to molecular signatures: EBV-positive (EBV), microsatellite instability (MSI), genomically stable (GS) and chromosomal instability (CIN) [1]. Many different signaling pathways, such as NF-κB and Hippo-YAP1 pathways, are aberrantly activated and involved in GC initiation and progression [2, 3]. In our previous study, we comprehensively summarized the oncogenic role of the Slit-Robo pathway in carcinogenesis [4]. However, the involvement of Slit-Robo pathway in gastric carcinogenesis remains poorly understood.
As the main binding partner and downstream effector of Slit-Robo signaling [5], SRGAP (Slit-Robo GTPase-activating protein) family was proposed to play a transduction role in the crosstalks of Slit-Robo and the other signaling pathways. This SRGAP family includes four members in mammals: SRGAP1, SRGAP2, SRGAP3 and ARHGAP4 [6], and these proteins have GAP activity for Rac [7,8,9], which indicates that SRGAPs regulate Rac activity. SRGAP consists of three functional domains, F-BAR, RhoGAP, SH3, and a functionally unknown carboxyl-terminus [6, 10]. The functional diversity of the membrane deformation properties is thought to be regulated by F-BAR domain and this domain is necessary for cell morphological changes [11]. SRGAPs interact with regulatory molecules of actin reorganization at the cell periphery, suggesting their involvement in the control of lamellipodial protrusions [12, 13]. In our previous study, we found that SRGAP1, but not SRGAP2 and SRGAP3, is significantly upregulated in GC. SRGAP1 is thought to play a crucial role in Slit-Robo signal transduction and its downstream signaling pathways. Most physiological studies focused on the regulation cell migration by SRGAP1 and found that SRGAP1 is a key determinant of lamellipodial dynamics and cell migratory behavior [14, 15]. However, how SRGAP1 upregulation and activation are involved in GC have not been elucidated. Thus, in this study, we will investigate what leads to upregulation of SRGAP1 and uncover how SRGAP1 activation promotes GC.
Results
SRGAP1 is upregulated in GC
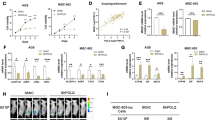
From the published GC data sets (NCBI/GEO/GSE27342 [16] and TCGA), SRGAP1, instead of SRGAP2 or SRGAP3, showed overexpression in GC samples compared with non-tumorous tissues (Fig. 1a and Supplementary Fig. 1a). Meanwhile, SRGAP1 was upregulated in 9 out 12 (75%) GC cell lines from both mRNA and protein level (Fig. 1b). In TCGA cohort, the mutation rate of SRGAP1 in primary GC is 3.1% (8/258). Additionally, SRGAP1 is amplified in 5/258 (1.9%) cases (Fig. 1c), and SRGAP1 mRNA expression is positively correlated with copy number change (Fig. 1d). However, among those patients with high SRGAP1 mRNA expression, 18 out of 258 cases are without gene amplification, indicating that alternative mechanisms are involved in its upregulation. To evaluate the expression pattern of SRGAP1 based on the molecular classification of GC, TCGA cohort was analyzed [17, 18]. SRGAP1 mRNA expression was enhanced in all four subtypes of GC when compared with normal gastric epithelium cells. Furthermore, SRGAP1 upregulation was mostly found in EBV-positive molecular subtype (Fig. 1e).
a SRGAP1 is upregulated in primary gastric tumors in both cohorts (P < 0.001, NCBI/GEO/GSE27342; P < 0.001, TCGA). b SRGAP1 is overexpressed in nine out of twelve GC cell lines in both mRNA and protein levels. c Genetic alteration rates of SRGAP1, Slit2, Robo1, and RhoA among GC patients in TCGA cohort. d SRGAP1 mRNA expression patterns within different genetic alterations in GC. e Expression pattern of SRGAP1 according to molecular classification. EBV EBV-positive; MSI microsatellite unstable; GS genomically stable; CIN chromosomal instability
SRGAP1 overexpression is correlated with poor prognosis of GC patients
SRGAP1 overexpression was correlated with poor overall and recurrence-free survival among GC patients in TCGA cohort (n = 378, P = 0.052; n = 350, P = 0.0895, Fig. 2a). When subgrouping the cohort according to Lauren Classification, upregulation of SRGAP1 significantly indicated a worse overall survival in intestinal type GC (n = 141, P = 0.0269, Fig. 2b). TCGA cohort was used to further elucidate the association between SRGAP1 expression and clinical parameters. Advanced stage and T/N stage were associated with SRGAP1 overexpression (Fig. 2c). On the other hand, gender, age, as well as H. pylori infection factors did not correlate with SRGAP1 abundance (Supplementary Fig. 1b–d). Moreover, gene set enrichment analysis (GSEA) on both the GC data sets revealed that cell metastasis-related genes were significantly enriched in SRGAP1 overexpressed cases (NCBI/GEO/GSE35809, P = 0.010, left panel; NCBI/GEO/GSE57303, P = 0.006, right panel; Fig. 2d) [19, 20]. Therefore, SRGAP1 might be critical for GC development through inducing cell metastasis. To elucidate the other potential mechanisms involved in SRGAP1 overexpression in GC, the methylation status of SRGAP1 was analyzed in TCGA database. Three CpG islands (cg05850206, cg22610533, cg14479829) were identified 600 bp ahead of the transcription start site (TSS) of SRGAP1. However, none of them contributed to the overexpression of SRGAP1 (Supplementary Fig. 1e).
a Upregulation of SRGAP1 indicated worse overall (left panel, P = 0.052) and recurrence-free survival (right panel, P = 0.0895) in TCGA cohort. b SRGAP1 abundance predicted poor recurrence-free survival in intestinal type GC (P = 0.0269). c Expression patterns of SRGAP1 based on Overall Stage Grouping (left panel; *P < 0.05), T stages (middle panel; *P < 0.05), and N stages (right panel; *P < 0.05). d Enrichment plots of gene expression signatures for metastasis according to SRGAP1 levels in two cohorts (NCBI/GEO/GSE35089, left panel, P = 0.010, n = 70; NCBI/GEO/GSE57303, right panel, P = 0.006, n = 70). The barcode plot indicated the position of the genes in each gene set; red and blue colors represented the high and low expression of SRGAP1, respectively. ES enrichment score; NES normalized enrichment score
SRGAP1 knockdown exerts tumor-suppressive effect and inhibits Wnt/β-catenin signaling
Given the abundance of SRGAP1 in GC, siRNA-mediated knockdown was performed in MKN28, MGC-803, and SGC-7901 cells for functional studies. A significantly decreased cell proliferation was detected in SRGAP1-depleted GC cells (P < 0.001, Fig. 3a). Also, SRGAP1 knockdown significantly reduced colony formation in these cell lines (P < 0.001, Fig. 3b and Supplementary Fig. 2a). In addition, siRNA-mediated knockdown of SRGAP1 decreased GC cell migration (P < 0.001, Fig. 3c and Supplementary Fig. 2b) and invasion (P < 0.001, Fig. 3d and Supplementary Fig. 2c). Since a growth inhibitory effect was observed in siSRGAP1-transfected cells, we analyzed the related cell cycle parameters using flow cytometry. However, there was no obvious alteration of cell cycle distribution after SRGAP1 knockdown (Supplementary Fig. 2d). Cell apoptosis, as determined by cleaved-Caspase 3 and cleaved-PARP, was validated by western blot in cells treated with siSRGAP1. Increased p21 and p27 expression was also detected in GC cell lines after SRGAP1 silencing, while MMP2 expression was suppressed (Fig. 3e). By Cancer 10-pathway Reporter Luciferase Kit, siSRGAP1 was found to significantly inhibit Wnt/β-catenin signaling in MGC-803 cells (Supplementary Fig. 2e). Then, the activity of Wnt/β-catenin signaling pathway was confirmed to be suppressed by SRGAP1 knockdown in MKN28, MGC-803 and SGC-7901 cell lines (Fig. 3f). We subsequently used TOPflash luciferase assays to validate that Wnt/β-catenin cascade was indeed inactivated by siSRGAP1 (Fig. 3g). Immunocytochemistry analysis revealed that siSRGAP1 inhibited the translocation of active-β-catenin and quenched its oncogenic role in the nucleus (Fig. 3h). Concordantly, suppression of Wnt/β-catenin signaling pathway by siSRGAP1 was evidenced by the decreased active-β-catenin, CCND1 and c-Myc expression in siSRGAP1-transfected GC cells (Fig. 3i).
a Six-day MTT assays revealed that SRGAP1 knockdown by siRNA significantly suppressed proliferation in GC cells (**P < 0.001). The mean and standard deviations (SDs) obtained from 6 wells were plotted. b Monolayer colony formation assays suggested that SRGAP1 depletion reduced anchorage-dependent colony formation (**P < 0.001). The experiments were done in triplicate and the error bars represented SDs. c SRGAP1 knockdown decreased cell migration ability of GC cells (** P < 0.001). Visions of cells were randomly selected for thrice, from which the SDs were achieved. d Cell invasion was significantly inhibited in SRGAP1-depleted GC cells (**P < 0.001). e Western blot analysis demonstrated the upregulation of cleaved-Caspase 3, p21/p27 and cleaved-PARP after SRGAP1 knockdown. f siSRGAP1 significantly inactivated Wnt/β-catenin signaling pathway in GC cells. Relative luciferase activity of Wnt/β-catenin signaling pathway in 3 gastric cell lines after siSRGAP1 treatment (left panel). Top/Flash reporter plasmid was transfected into the SRGAP1 knockdown MKN28, MGC-803 and SGC-7901 cells, the reporter activities were determined by luciferase assay (right panel; **P < 0.001). g The effect of siSRGAP1 on the active-β-catenin was analyzed by immunocytochemistry staining with anti-active-β-catenin antibody (Red) and DAPI nuclear staining (blue). h Western blot of active-β-catenin, β-catenin, CCND1/3 and c-Myc after knocking down SRGAP1 in MKN28, MGC-803 and SGC-7901 cells. i siSRGAP1 suppressed Wnt/β-catenin signaling pathway in GC cells
siSRGAP1 induces accumulation of GTP-RhoA and GTP-Cdc42 and inhibits EMT (epithelial-mesenchymal transition) in GC
Polymerization and depolymerization of filamentous actin (F-actin) control cytoskeletal reorganization, leading to morphologic changes related to cell motility. Cellular morphological changes were observed after siSRGAP1 transfection, with a significantly increased quantity of cell protrusions. Therefore, the capability of SRGAP1 in regard to regulating F-actin distribution was investigated by F-actin staining with FITC-conjugated phalloidin. Results from immunofluorescence analysis showed that stress fibers in siScramble-treated cells displayed a well-organized manner; however, the F-actin was loosely organized in the cells after knocking down SRGAP1. In addition, the number of cell protrusions was significantly increased in siSRGAP1 transfected SGC-7901 (Fig. 4a), which could partly explain its inhibitory effect on cell migration and invasion. GAPs inactivate Rho proteins by stimulating their GTPase activity and SRGAP1 can promote the GTP hydrolysis of RhoA, Cdc42 or Rac [21]. To determine whether SRGAP1 affects the GTP/GDP-binding status of Rho GTPases, GTP-bound RhoA, Rac1 and Cdc42 were chosen for verification through pull-down assays in SGC-7901 cells. The results showed that siSRGAP1 increased the accumulation of GTP-RhoA and GTP-Cdc42, while promoted GTP-Rac1 hydrolysis. Hence, suppression of SRGAP1 affected the amount of active GTP-bound RhoA, Rac1 and Cdc42, thus altered cell morphology and inhibited cell migration (Fig. 4b). Accumulating evidence highlighted EMT as the mechanism through which epithelial cancers achieve motile and invasive capabilities to progress toward a more aggressive phenotype. By immunofluorescence analysis, SRGAP1 knockdown led to decreased expression of N-cadherin and Vimentin compared with the control group (Fig. 4c). To further confirm the EMT phenotype, both N-cadherin and Vimentin showed decreased expression by western blot analysis (Fig. 4d). Consistently, in TCGA cohort, SRGAP1 mRNA expression exhibited negative correlation with RhoA mRNA expression (r = −0.291, n = 415, P < 0.001, Fig. 4e). For EMT markers, mRNA expression of N-cadherin (r = 0.120, n = 415, P = 0.015, Fig. 4e), instead of Vimentin (r = 0.043, n = 415, P = 0.383, Fig. 4e), displayed a positive association with SRGAP1 mRNA expression, indicating that SRGAP1 induced EMT through N-cadherin in vivo. Moreover, the oncogenic role of SRGAP1 was validated in vivo. SRGAP1 overexpression significantly enhanced the growth of tumor xenografts in nude mice (P < 0.001, Fig. 4f).
a FITC-phalloidin staining of F-actin in siScramble and siSRGAP1 cells (left panel). 100 cells for siScramble and siSRGAP1, respectively, were assessed across three independent experiments. And quantification of cell protrusions in cells treated with siSRGAP1 was shown in the right panel. b The effect of siSRGAP1 on GTPase guanine nucleotide-binding status of RhoA, Rac1 and Cdc42. A significant increase of GTP-RhoA and GTP-Cdc42 was detected, while GTP-Rac1 level was decreased after SRGAP1 knockdown. c siScramble and siSRGAP1 cells were fluorescence-stained for N-cadherin (green), Vimentin (green), and DAPI (blue) respectively. d Protein expression of N-cadherin and Vimentin was inhibited after SRGAP1 silencing in GC cell lines. e Expression correlation between SRGAP1 mRNA expression and RhoA (left panel, r = −0.291, P < 0.001), N-cadherin (middle panel, r = 0.120, P = 0.015) and Vimentin (right panel, r = 0.043, P = 0.383) mRNA expression respectively according to TCGA cohort. f SRGAP1 overexpression promotes xenograft formation in nude mice (P < 0.001)
To further explore the possible mechanisms that how SRGAP1 is involved in cell migration and invasion, RNA Sequencing (RNA-Seq) analysis was performed in siScramble or siSRGAP1 treated GC cells. Three significantly dysregulated gene sets (WU_CELL_MIGRATION, CHANG_CYCLING_GENES, and GABRIELY_MIR21-TARGETS) related to cell migration or invasion have been identified (Supplementary Fig. 3a). The heatmap demonstrated the differentially expressed genes in these three gene sets, respectively (Supplementary Fig. 3b and Supplementary Tables 1–3. GESA revealed that SRGAP1 knockdown was negatively correlated with the enriched signaling pathways (Supplementary Fig. 3c). To validate the RNA-Seq data, eight candidate genes (AKR1C2, AKR1C1, AKR1C3, HMGA2, RGS20, CYR61, FGF2, and WNT5A) were selected. These genes are crucial mediators that regulate cell migration and invasion. All designed primers were listed in Supplementary Fig. 3d. Low expressions of all these eight genes in the siSRGAP1 group were detected by qRT-PCR (Supplementary Fig. 3e), indicating that SRGAP1 regulates the expression of downstream thus to promote tumor progression by enhancing cell migration and invasion.
SRGAP1 is negatively regulated by miR-340 and miR-124 in GC
By bioinformatics analysis (http://www.microrna.org/microrna/home.do), we screened out top-20 miRNAs that might regulate SRGAP1 according to the SVR score (Supplementary Table 4). Furthermore, with TargetScan for the secondary screening (www.targetscan.org), miR-340 and miR-124 were noticed as they have multiple putative targets on SRGAP1 3’UTR (three binding sites for miR-340 and miR-124 in its 3′-UTR, respectively) (Fig. 5a and b). In GC cell lines, miR-340 or miR-124 reduce SRGAP1 expression in both mRNA and protein levels (P < 0.001, Fig. 5c and d). Moreover, luciferase assays were performed to validate the direct binding affinity between SRGAP1 3′UTR and these two miRNAs. The fragments of SRGAP1 3′UTR containing the predicted or mutant miR-340 and miR-124 binding sites 1, 2 and 3 were subcloned into pMIR-REPORT vectors. For miR-340, we found that ectopic expression exerted a significant inhibitory effect on the luciferase activity in the constructs that contain the wild-type sequence of binding site 1 and 3, whereas no suppressive effects were observed for the construct of binding site 2 (left panel, Fig. 5e). Similarly, luciferase activity was also decreased after ectopic expression of miR-124 in the constructs, which consist of the wild-type sequence of binding site 2 and 3 (right panel, Fig. 5e). These results revealed that miR-340 and miR-124 specifically and directly suppressed SRGAP1 expression by binding to its 3′-UTRs.
a The top 6 highest mirSVR scores of microRNAs that target SRGAP1 according to a microRNA database TargetScan (www.targetscan.org). b Predicted putative miR-340 and miR-124-binding sites in 3’-UTR of SRGAP1. c Both mRNA and protein expression of SRGAP1 after miR-340 overexpression in MKN28, MGC-803 and SGC-7901 cells (**P < 0.001). d miR-124 decreased the expression of SRGAP1 in GC cells both from mRNA and protein levels (**P < 0.001). e miR-340 suppressed the relative luciferase activity in the constructs that contain the binding site 1 and 3 in the 3’-UTR of SRGAP1, whereas miR-124 inhibited the ones possessing the binding sites 2 and 3 (**P < 0.001)
Furthermore, we found that miR-340 and miR-124 regulated cell-invasion-related genes according to miRBase prediction (Supplementary Fig. 4a, c). Both mRNA and protein levels of RhoA, ROCK1, and MET were negatively modulated by miR-340 (Supplementary Fig. 4b). Similarly, miR-124 downregulated RhoG, ROCK1 and CTGF in GC cell lines (Supplementary Fig. 4d).
miR-340 and miR-124 are downregulated and display anti-tumor effect in GC
Expression of miR-340 and miR-124 were downregulated in multiple GC cell lines compared with normal control (GES-1) (Fig. 6a). After treating with 5-Aza, TSA or the combination of both, miR-340 and miR-124 showed significantly increased expression, suggesting that promoter methylation or histone deacetylation may be responsible for the downregulation of these miRNAs in GC (Fig. 6b). To confirm whether miR-340 and miR-124 were downregulated via promoter hypermethylation, the methylation status of miR-340 and miR-124 were evaluated by bisulfite genome sequencing (BGS) analysis. BGS primers were designed using MethPrimer 2.0 (Supplementary Fig. 5a). For miR-124, GC cell lines (MKN45, MGC-803, and NCI-N87) exhibited dense DNA methylation, whereas immortalized normal gastric epithelial cell line (GES1) showed a low degree of methylation at each CpG site evaluated (Supplementary Fig. 5b). Similarly, the methylation of miR-340 promoter region was also evaluated by BGS analysis. A high level of CpG methylation was detected in NCI-N87, MKN45, and MGC-803, but no CpG site methylation was observed in GES1 (Supplementary Fig. 5c).
a Expression of miR-340 and miR-124 was downregulated in most of GC cell lines comparing with normal gastric epithelium tissue. b Expression of miR-340 and miR-124 was restored in NCI-N87, MKN1 and MGC-803 cells after treated with 5-Aza and TSA. c six-day MTT proliferation results with ectopic miR-340 and miR-124 expression in GC cells (**P < 0.001). The error bars represented SDs. d Ectopic expression of miR-340 and miR-124 inhibited monolayer colony formation in MKN28, MGC-803 and SGC-7901 cells (**P < 0.001). The experiment was performed in triplicate wells to get SDs. e The invasive ability was significantly impaired in miR-340- and miR-124-treated cells comparing with scramble miRNA counterparts (**P < 0.001). f Flow cytometry analysis of miR-340 and miR-124 and scramble miRNA transfectants. The representative bar chart of cell distribution indicated the same trend of two independent experiments. g Ectopic expression of miR-340 and miR-124 induced senescence (**P < 0.001). SDs were determined by the positive cell number that counted from three randomly-picked fields. h Western blot of cell cycle regulators with miR-340 and miR-124 highly expressed
Given that miR-340 and miR-124 are often downregulated in GC, ectopic expression of miR-340 and miR-124 was applied to identify their functional roles. Overexpressed miR-340 and miR-124 suppressed GC cell growth accordingly (P < 0.001, Fig. 6c). Moreover, a significant reduction of colony numbers in miRNAs’ transfectants was detected compared with scramble miRNA groups (P < 0.001, Fig. 6d and Supplementary Fig. 6a). In addition, ectopic expression of miR-340 and miR-124 significantly diminished cell invasion (P < 0.001, Fig. 6e and Supplementary Fig. 6b). Given the inhibitory effect of miR-340 and miR-124 on GC cell proliferation, we next investigated their possible underlying mechanisms. Ectopic expression of miR-340 and miR-124 promoted G1 cell cycle arrest 24 h after transfection (MKN28, from 67.1 to 74.6% or 74%; MGC-803, from 60.5 to 66.7% or 76.4%; SGC-7901, from 63.7 to 66.1% or 73.3%), and the percentage of S-phase cells decreased accordingly (Fig. 6f and Supplementary Fig. 6c). Moreover, miR-340 and miR-124 transfectants also induced senescence (P < 0.001, Fig. 6g and Supplementary Fig. 6d), concordant with the G1-phase cell cycle arrest. Western blot analysis suggested a decrease of phosphorylation of retinoblastoma protein (p-Rb) and elevated expression of p21 and p27 in both miR-340- and miR-124-treated groups, in keeping with the results above (Fig. 6h). In addition, miR-340 and miR-124 induced late apoptosis in GC cells, further suggesting their tumor-suppressive role in GC (Supplementary Fig. 6e).
Downregulation of miR-340 serves as an independent prognostic marker and enhances drug sensitivity in GC
In 76 paired primary RNA samples, miR-340 and miR-124 were found to be downregulated in 52 (68.4%) and 51 (67.1%) tumor tissues compared to adjacent normal gastric tissue respectively (n = 76, P < 0.001, Fig. 7a, b). In this cohort, for each miRNA, the low expression group was always associated with worse survival (miR-340, P = 0.004, Fig. 7c; miR-124, P = 0.048, Fig. 7d). Consistently, in TCGA data set, reduction of miR-340 was also correlated with poor clinical outcomes (overall survival, n = 400, P = 0.024, left panel; recurrence-free survival, n = 316, P = 0.038, right panel, Fig. 7e). Then, the correlation between clinicopathologic parameters (gender, advanced age, intestinal or diffuse type, TNM stage, lymph node metastasis, H. pylori infection) and miR-340/124 was also checked. However, no statistically significant result was found (Supplementary Table 5). Furthermore, the data from both univariate and multivariate Cox regression analysis indicated that only advanced T-stage and low expression of miR-340 were associated with poor prognosis independently (T-stage, P = 0.001; miR-340, P = 0.025; Supplementary Table 6). To further evaluate the application of the potential therapeutic treatment, drug sensitivity experiments were performed. In MGC-803 and SGC-7901 cells, both SRGAP1 knockdown and miR-340/124 overexpression reduced cell viability when treated with chemotherapeutic agents (5-FU and Cisplatin, Fig. 7f).
a miR-340 expression is suppressed in gastric tumor tissues in contrast to adjacent non-tumorous tissues (n = 76, P < 0.001). b Downregulation of miR-124 was detected in primary samples (n = 76, P < 0.001). c Low miR-340 expression indicates poor recurrence-free survival (P = 0.004, n = 76). d miR-124 downregulation is related to worse recurrence-free outcome (P = 0.048, n = 76). e Low level of miR-340 indicated both unfavorable overall (left panel, P = 0.024, n = 400) and recurrence survival (right panel, P = 0.038, n = 316) according to TCGA cohort. f SRGAP1 knockdown or miR-340/miR-124 overexpression enhanced drug sensitivity of GC cells. The viability of treated cells was monitored for 48 h using MTT assay in MGC-803 and SGC-7901 cell lines with different dose of 5-FU and Cisplatin, respectively
SRGAP1 is the bona fide functional target of miR-340 in gastric carcinogenesis
Rescue experiments were also conducted to justify whether SRGAP1 is the functional target of miR-340. SRGAP1 overexpression was confirmed by western blot analysis (Fig. 8a). The re-expression of SRGAP1 partly diminished the tumor-suppressive effect of miR-340 (MTT proliferation assays, P < 0.001, Fig. 8b; monolayer colony formation, P < 0.001, Fig. 8c; cell invasion, P < 0.001, Fig. 8d). In primary gastric tumors from the TCGA cohort, SRGAP1 mRNA was negatively correlated with miR-340 expression (r = −0.131, n = 412, P = 0.008, Fig. 8e). Hereto, a piece of regulatory network involving SRGAP1 was elucidated. SRGAP1, a downstream effector of Slit-Robo pathway, functions as an oncogene through activating Wnt/β-catenin pathway. It also modulates cell migration by promoting hydrolysis of GTP-RhoA and GTP-Cdc42. The activation of SRGAP1 is partly due to dysregulation of miR-340 and miR-124 in GC. Our results highlight SRGAP1 as a link of signal transduction between Slit-Robo pathway and Wnt/β-catenin pathway (Fig. 8f).
a Re-overexpression of SRGAP1 in protein level among miR-340-treated GC cell lines. b Overexpressed SRGAP1 partially diminished the tumor-suppressive effect of miR-340 by 5-day MTT proliferation assays (*P < 0.05; **P < 0.001). c Colony formation was partly restored in miR-340 transfectants after re-expression of SRGAP1 (**P < 0.001). d SRGAP1 re-expression also revived cell invasive ability which was impaired by miR-340 (**P < 0.001). e miR-340 is negatively correlated with SRGAP1 expression in GC (r = −0.131, P = 0.008, n = 412). f The schematic representation of SRGAP1 as a linker of Slit-Robo pathway and Wnt/β-catenin pathway. SRGAP1, a downstream component of Slit-Robo cascade, exerts its oncogenic function through Wnt/β-catenin signaling. It also regulates cell migration by hydrolysis of GTP-RhoA and GTP-Cdc42. Moreover, its activation is in part attributed to the epigenetic silence of miR-340 and miR-124 in GC
Discussion
Our study for the first time reported the oncogenic role of SRGAP1 in tumorigenesis, especially in GC. Previously, we proposed that SRGAP1 might have some effect in the crosstalk between Slit-Robo and Wnt/β-catenin pathway [4]. In colorectal cancer, blocked Slit-Robo prohibited E-cadherin degradation and led to EMT reversion, exhibiting an ability of anti-tumor and anti-metastasis [22]. Via Wnt/β-catenin kinases, Slit-Robo also facilitated tumor growth in intestinal malignancies [23]. In this study, we identified SRGAP1 as the direct link between Slit-Robo and Wnt/β-catenin signal transduction and provided the first evidence that Slit-Robo signaling promotes tumorigenesis through SRGAP1.
As a GTPase activation protein, the main physiological function of SRGAP1 is to hydrolyze Rho-GTP to Rho-GDP [24]. Rho family GTPases are known to regulate a variety of cellular processes, including cell migration. The three best-characterized Rho family members are Cdc42, RhoA and Rac1. These small GTPases are essential in the regulation of actin cytoskeletal dynamics to influence cell migration [25]. Rho GTPases have been suggested to promote tumorigenesis and serves as useful anti-neoplastic targets for over two decades. However, several recent mouse model studies have pointed out that Rac1, RhoA and some of their downstream effectors, can also exhibit a tumor-suppressive role in defined condition [26]. RhoA activation increases the stress fiber and focal adhesion complexes. Cdc42 activation boosts filopodia formation and generates large, multinucleated cells that enhance anchorage-independent cell growth. Rac1 activation enhances the formation of lamellipodia [27]. Activated and inactivated by guanine nucleotide-exchange factors (GEFs) and GAPs respectively, these small GTPases are altered between GDP-form and GTP-form. Here, our findings revealed that SRGAP1 critically regulates RhoA, Cdc42 and Rac1 activity. Interacting with Slit ligands changes the domains of Robo receptors within the cytosol, inducing the binding between SRGAP1 and Robo1’s intracellular domain, by which Cdc42 is silenced while RhoA and Rac are modestly activated [21, 28]. It has been suggested that SRGAP1 binds to CDC42 and RhoA, that SRGAP2 and SRGAP3 interact with Rac1 via RhoGAP domain to promote GTP hydrolysis [29]. Our results indicate that Slit-Robo signaling dynamically regulates rearrangements of the actin cytoskeleton through SRGAP1 [30]. Furthermore, our results provide novel evidence that SRGAP1 promotes metastasis in GC, which may be mediated through the RhoA and Cdc42-related pathways. Apart from the direct hydrolysis on Rho family by SRGAP1, multiple genes (AKR1C2, AKR1C1, AKR1C3, HMGA2, RGS20, CYR61, FGF2, and WNT5A) associated with cell migration and invasion showed decreased expression upon SRGAP1 knockdown revealed by RNA-Seq. Therefore, in addition to direct hydrolyzing Rho GTPases, SRGAP1 also regulates the mRNA expression of some migration/invasion-related genes to exert its oncogenic function. Our findings revealed two independent mechanisms that SRGAP1 were involved in cell migration and invasion in gastric carcinogenesis.
As some GC cases show SRGAP1 mRNA upregulation without gene amplification, miRNA regulation on SRGAP1 was further investigated. We screened out miR-340 and miR-124 as two promising miRNAs potentially targeting SRGAP1 in GC. The activation of SRGAP1 is partly due to epigenetic silencing of these two miRNAs. It has been reported that miR-340 is silenced in cancers and it exerts tumor-suppressive function in breast cancer [31], osteosarcoma [32], melanoma [33], non-small cell lung cancer [34] and gliomas [35]. In hepatocellular carcinoma (HCC) cells, miR-340 antagonizes cisplatin resistance by targeting antioxidant pathway in a Nrf2-dependent manner [36]. miR-124 has also been confirmed as a tumor-suppressive miRNA in multiple cancer types, such as bladder cancer [37], nasopharyngeal carcinoma [38], breast cancer [39], glioma [40] and HCC [41]. In primary GC samples, the expression of miR-340 exhibits negative correlation with SRGAP1 and the re-expression of SRGAP1 partly abolished the growth inhibitory effect of miR-340, suggesting SRGAP1 is a key target of miR-340 in gastric carcinogenesis. Our study enriched the target pool for miR-340 and miR-124 in GC. The aberrant regulation of SRGAP1 by this dysregulated miRNA network is novel. In previous studies, SRGAP1 was found to have a relatively high mutation rate in cancers. In ovarian cancer, a risk genetic variant in SRGAP1 (rs11175194) was revealed by genome-wide association study (GWAS) [42]. The SRGAP1 missense variants, Q149H, A275T and R617C, were uncovered in papillary thyroid carcinoma and proposed to impair its Cdc42 inhibition ability [43]. Herein, we found that the aberrant upregulation of SRGAP1, mediated by the epigenetic silence of two miRNAs, can also contribute to GC development in the absence of genetic mutations.
In conclusion, SRGAP1 functions as an oncogene in gastric tumorigenesis especially by promoting metastasis. Multiple mechanisms contribute to SRGAP1 upregulation and activation, including gene amplification, gain-of-function mutation, and miRNA regulation. Our study provided a novel therapeutic target and generated the clinical translational potential for GC.
Materials and methods
GC cell lines and primary tissues
GC cell lines used in this study have been reported [44], primary tumors have also been described [45]. Ethical approval was obtained from the joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (CREC Ref. No.2015.134) has approved the ethical permission.
5-Aza and TSA treatment
Demethylation agent (5-Aza) and histone deacetylases inhibitor were described previously [46, 47]. Treatment procedures have been indicated in our previous work [48].
Bisulphite genomic sequencing
miR-340 and miR-124 CpG island methylation were analyzed by bisulphite genomic sequencing. Briefly, 1 μg genomic DNA extracted from GES1, MKN45, MGC-803, and NCI-N87 was treated with sodium bisulfite. The converted DNA was amplified using 1 μl template, 10 × PCR buffer, 0.2 μM of each primer, 250 μM dNTP mix and 1 μl Taq polymerase. PCR products were purified and then sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and analyzed using an Applied Biosystems 3130 Genetic Analyzer.
RNA extraction and qRT-PCR
Total RNA extraction was illustrated previously [49]. All the primes were listed in Supplementary Table 7. miRNA expression detection was also performed accordingly [45].
miRNA/ siRNA transfection
The miRNA precursors miR-340 (PM12670), miR-124 (PM10691) were from Life Technologies, and siSRGAP1 (SI00733026) was from Qiagen (Valencia, CA, USA). The other reagents were same as before [46]. Detailed protocols were as described previously [46].
Western blot
Antibodies from cell signaling have been indicated before [49, 50]. Other antibodies included anti-SRGAP1 antibody (1:1000, sc-81939, Santa Cruz Biotechnology, Dallas, TX, USA), RhoA (#05-778), Cdc42 (#05-542), Rac1 (#05-389), N-cadherin (33-3900, Invitrogen), and Vimentin (YM3158, ImmunoWay Biotechnology Company, Plano, TX, USA). The secondary antibodies were in line with that in our earlier work [49].
Immunofluorescence imaging
For immunofluorescence, SGC-7901 cells were transfected with siScramble or siSRGAP1 for 48 h, then washed with cold PBS, and fixed with 4% PFA (Sigma-Aldrich). Non-specific binding was blocked with normal goat serum (Invitrogen). Primary antibodies N-cadherin (1:100, 33-3900, Invitrogen), ABC (1:100, #05-665, Millipore, Billerica, MA, USA) and Vimentin (1:100, YM3158, ImmunoWay) were applied at 4 °C overnight. The secondary antibody AlexaFluor (1:500, Invitrogen) was added at RT for 1 h and the cell nuclei were counterstained with DAPI (Sigma-Aldrich). For F-actin staining, FITC-labeled phalloidin (P5282, Sigma-Aldrich) was applied to incubate at RT for 40 min and the cell nuclei counterstained with DAPI. Images were captured with a confocal microscope Carl Zeiss LSM880 (Carl Zeiss, Gottingen, Germany).
Pathway analysis
The procedures were indicated in our earlier paper [50].
GTPase activity assays
siScramble or siSRGAP1-transfected SGC-7901 cells lysed in Mg2+ lysis buffer were assayed using the Rho activation assay kit (#17-294, Upstate, Millipore, Bedford, MA, USA) or Rac1/Cdc42 activation assay kit (#17-441). The buffer was supplemented with 10% glycerol. The lysed cells were centrifuged and the supernatant collected was added to 20 μl slurry of Rho Assay of Rho Assay reagent (Rhotekin Rho binding domain) or 10 μl slurry of Rac1/Cdc42 assay reagent (PAK-1 PBD, agarose) according to the manufacturer’s instructions. The reaction mixture was gently mixed and the agarose beads collected by centrifugation. After washing, re-suspended beads were boiled with SDS-PAGE. Western blots were performed and the amounts of GTP-bound RhoA, Rac1 and Cdc42 were compared to their total amounts in the whole-cell crude lysate.
RNA-Seq analysis
RNA from siScramble or siSRGAP1 transfected SGC-7901 cells was extracted by using the RNeasy kit (Qiagen, Valencia, CA, USA). Quality control for RNA samples was performed by the Humanizing Genomics Macrogene Company (Seoul, Republic of Korea). Then, samples were prepared using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA). After the preparation, samples were sequenced on the Illumina Hiseq 4000 100PE system and analyzed. GSEA was performed to identify differentially expressed gene sets (with a Benjamini & Hochberg-adjusted P value <0.05) related to cell migration or invasion.
5-Fluorouracil and Cisplatin treatment
The cells were seeded onto 96-well plates and incubated with 10% FBS at 37 °C. After 24 h, RPMI 1640 containing various concentration of 5-Fluorouracil (5-FU) (0, 10, 20, 50, 100, 250 μg/ml) or Cisplatin (0, 4, 8, 12, 16, 20 μg/ml) was added to each well. The cellular viability was quantified by the 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) assay after another 24 h. All MTT assays include triplicate wells for each time point of each cell line.
Luciferase assays
For reporter assay, sense and antisense oligonucleotides were illustrated in Supplementary Table 8, cells were transiently co-transfected with reporter plasmid and miR-340, miR-124 or control-miRNA. Measurement has also been described [46].
Tumorigenicity model for animals
Protocols were demonstrated in an earlier time [49, 50]. All operations were approved by the Animal Ethics Committee of the Chinese University of Hong Kong.
Statistical analysis
Some values were adjusted (log transformation) to reach the requirements (normally distributed or approximately normally distributed) of parametric tests. Paired t test was employed to compare SRGAP1 knockdown cells and scramble siRNA-transfected counterparts. Other detailed applications of statistical methods were demonstrated as before [48].
References
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Huang T, Kang W, Zhang B, Wu F, Dong Y, Tong JHM, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-κB signaling in gastric carcinogenesis. Mol Cancer. 2016;15:9.
Kang W, Cheng ASL, Yu J, To KF. Emerging role of Hippo pathway in gastric and other gastrointestinal cancers. World J Gastroenterol. 2016;22:1279.
Huang T, Kang W, Cheng ASL, Yu J, To KF. The emerging role of Slit-Robo pathway in gastric and other gastro intestinal cancers. BMC Cancer. 2015;15:950.
Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cel.l 2001;107:209–21.
Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86.
Yang Y, Marcello M, Endris V, Saffrich R, Fischer R, Trendelenburg MF, et al. MEGAP impedes cell migration via regulating actin and microtubule dynamics and focal complex formation. Exp Cell Res. 2006;312:2379–93.
Mason FM, Heimsath EG, Higgs HN, Soderling SH. Bi-modal regulation of a formin by srGAP2. J Biol Chem. 2011;286:6577–86.
Neukomm LJ, Frei AP, Cabello J, Kinchen JM, Zaidel-Bar R, Ma Z, et al. Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat Cell Biol. 2011;13:79–86.
Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804.
Coutinho-Budd J, Ghukasyan V, Zylka MJ, Polleux F. The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J Cell Sci. 2012;125:3390–401.
Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, et al. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38.
Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T, et al. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat Cell Biol. 2002;4:970–5.
Yamazaki D, Itoh T, Miki H, Takenawa T. srGAP1 regulates lamellipodial dynamics and cell migratory behavior by modulating Rac1 activity. Mol Biol Cell. 2013;24:3393–405.
Kutys ML, Yamada KM. An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat Cell Biol. 2014;16:909–17.
Cui J, Chen Y, Chou W-C, Sun L, Chen L, Suo J, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–207.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1–pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Chia N-Y, Deng N, Das K, Huang D, Hu L, Zhu Y, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–19.
Qian Z, Zhu G, Tang L, Wang M, Zhang L, Fu J, et al. Whole genome gene copy number profiling of gastric cancer identifies PAK1 and KRAS gene amplification as therapy targets. Genes Chromosomes Cancer. 2014;53:883–94.
Wong K, Ren X-R, Huang Y-Z, Xie Y, Liu G, Saito H, et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the slit-robo pathway. Cell. 2001;107:209–21.
Zhou WJ, Geng ZH, Chi S, Zhang W, Niu XF, Lan SJ, et al. Slit-Robo signaling induces malignant transformation through Hakai-mediated E-cadherin degradation during colorectal epithelial cell carcinogenesis. Cell Res. 2011;21:609–26.
Zhang QQ, Zhou DL, Lei Y, Zheng L, Chen SX, Gou HJ, et al. Slit2/Robo1 signaling promotes intestinal tumorigenesis through Src-mediated activation of the Wnt/beta-catenin pathway. Oncotarget. 2015;6:3123–35.
Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35.
Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701.
Zandvakili I, Lin Y, Morris JC, Zheng Y. Rho GTPases: Anti-or pro-neoplastic targets? Oncogene. 2017;36:3213.
Lin R, Cerione RA, Manor D. Specific Contributions of the Small GTPases Rho, Rac, and Cdc42 to Dbl Transformation. J Biol Chem. 1999;274:23633–41.
Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–5.
Liu S, Xiong X, Zhao X, Yang X, Wang H. F-BAR family proteins, emerging regulators for cell membrane dynamic changes—from structure to human diseases. J Hematol Oncol. 2015;8:1–14.
Ypsilanti AR, Zagar Y, Chédotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–52.
Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, et al. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer. 2011;117:2842–52.
Zhou X, Wei M, Wang W. MicroRNA-340 suppresses osteosarcoma tumor growth and metastasis by directly targeting ROCK1. Biochem Biophys Res Commun. 2013;437:653–8.
Poenitzsch Strong AM, Setaluri V, Spiegelman VS. MicroRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma. Arch Biochem Biophys. 2014;563:118–24.
Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2014;34:3240–50.
Yamashita D, Kondo T, Ohue S, Takahashi H, Ishikawa M, Matoba R, et al. miR-340 suppresses the stem-like cell function of glioma-initiating cells by targeting tissue plasminogen activator. Cancer Res. 2015;75:1123–33.
Shi L, Chen ZG, Wu LL, Zheng JJ, Yang JR, Chen XF, et al. miR-340 Reverses Cisplatin Resistance of Hepatocellular Carcinoma Cell Lines by Targeting Nrf2-dependent Antioxidant Pathway. Asian Pac J Cancer Prev. 2014;15:10439–44.
Zhang T, Wang J, Zhai X, Li H, Li C, Chang J. MiR-124 retards bladder cancer growth by directly targeting CDK4. Acta Biochim Biophys Sin. 2014;46:1072–9.
Peng XH, Huang HR, Lu J, Liu X, Zhao FP, Zhang B, et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol Cancer. 2014;13:186.
Li W, Zang W, Liu P, Wang Y, Du Y, Chen X, et al. MicroRNA-124 inhibits cellular proliferation and invasion by targeting Ets-1 in breast cancer. Tumour Biol. 2014;35:10897–904.
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 2014;16:1341–53.
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–89.
Chen K, Ma H, Li L, Zang R, Wang C, Song F, et al. Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nat Commun. 2014;5:4682.
He H, Bronisz A, Liyanarachchi S, Nagy R, Li W, Huang Y, et al. SRGAP1 is a candidate gene for papillary thyroid carcinoma susceptibility. J Clin Endocrinol Metab. 2013;98:E973–980.
Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–9.
Kang W, Tong JH, Lung RW, Dong Y, Yang W, Pan Y, et al. let-7b/g silencing activates AKT signaling to promote gastric carcinogenesis. J Transl Med. 2014;12:281.
Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14:52.
Zehentmayr F, Hauser-Kronberger C, Zellinger B, Hlubek F, Schuster C, Bodenhofer U, et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin Epigenetics. 2016;8:28.
Zhou Y, Huang T, Zhang J, Wong CC, Zhang B, Dong Y et al. TEAD1/4 exerts oncogenic role and is negatively regulated by miR-4269 in gastric tumorigenesis. Oncogene. 2017;36:6518–6530.
Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017;16:77.
Kang W, Tong JHM, Chan AWH, Zhao J, Dong Y, Wang S, et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J Transl Med. 2014;12:80.
Acknowledgements
We acknowledge the TCGA Research Network (http://cancergenome.nih.gov/), The UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/), and NCI Center for Cancer Genomics Office (http://gdc.nci.nih.gov/) for providing the gastric cancer data set and analysis. This study is supported by General Research Fund (RGC Reference No. CUHK14114414 and CUHK14110016) from The Research Grants Council of Hong Kong. We also acknowledge the technical support from Core Utilities of Cancer Genomics and Pathobiology of Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong.
Author contributions
KFT and WK: Designed the experiments, offered direction and help on the whole project. TH, YZ, JZ, CCW, WL, JSHK, RY, AKYC, YD, FW, BZ, and AHKC: Conducted the experiments, analyzed the results and performed bioinformatics analysis. TH, YZ, and WK: Drafted the manuscript. WKKW, ASLC, JY, and NW: Reviewed the manuscript and made significant revisions on the drafts. All authors participated in the data interpretation and critically revised and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Tingting Huang and Yuhang Zhou contributed equally to this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, T., Zhou, Y., Zhang, J. et al. SRGAP1, a crucial target of miR-340 and miR-124, functions as a potential oncogene in gastric tumorigenesis. Oncogene 37, 1159–1174 (2018). https://doi.org/10.1038/s41388-017-0029-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-017-0029-7
This article is cited by
-
LncRNA SND1-IT1 facilitates TGF-β1-induced epithelial-to-mesenchymal transition via miR-124/COL4A1 axis in gastric cancer
Cell Death Discovery (2022)
-
The emerging roles of srGAPs in cancer
Molecular Biology Reports (2022)
-
Overexpression of miR-340 inhibits cell proliferation and induces apoptosis of human bladder cancer via targeting Glut-1
BMC Urology (2021)
-
Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340
Molecular Cancer (2020)
-
miR-340-FHL2 axis inhibits cell growth and metastasis in ovarian cancer
Cell Death & Disease (2019)