Abstract
Pancreatic acinar cell carcinoma is relatively rare (1 to 2% of pancreatic malignancies) but may be under-recognized. In contrast to pancreatic ductal adenocarcinoma, most acinar cell carcinomas lack mutations in KRAS, DPC, CDKN2A or TP53, but appear to have a high incidence of gene rearrangements, with up to 20% reported to be driven by BRAF fusions. With the development of a new class of RET-specific tyrosine kinase inhibitors, which appear to have particularly strong activity against RET gene rearranged tumours, there is now considerable interest in identifying RET gene rearrangements across a wide range of cancers. RET rearrangements have been reported to occur at a very low incidence (<1%) in all pancreatic carcinomas. We postulated that given its unique molecular profile, RET gene rearrangements may be common in acinar cell carcinomas. We performed fluorescent in-situ hybridization (FISH) studies on a cohort of 40 acinar cell spectrum tumours comprising 36 pure acinar cell carcinomas, three pancreatoblastomas and one mixed acinar-pancreatic neuroendocrine tumour. RET gene rearrangements were identified in 3 (7.5%) cases and BRAF gene rearrangements in 5 (12.5%). All gene rearranged tumours were pure acinar cell carcinomas. Our findings indicate that amongst all pancreatic carcinomas, acinar carcinomas are highly enriched for potentially actionable gene rearrangements in RET or BRAF. FISH testing is inexpensive and readily available in the routine clinical setting and may have a role in the assessment of all acinar cell carcinomas—at this stage to recruit patients for clinical trials of new targeted therapies, but perhaps in the near future as part of routine care.
Similar content being viewed by others
Introduction
Pancreatic acinar cell carcinoma is a relatively rare neoplasm which accounts for less than 1–2% of pancreatic malignancies in adults and ~15% of pancreatic malignancies in children where there is some morphological and clinical overlap with pancreatoblastoma (differentiated from acinar cell carcinoma by the presence of squamous morules) [1,2,3]. Early reports suggested a 3-year survival as low as 26% and a mean survival of 18 months with ~50% of patients presenting with metastatic disease [1, 4, 5]. More recent data have indicated an improved 5-year survival of 43% (up to 72% if undergoing resection; 22% for those who are inoperable) – summarized by Klimstra et al. and La Rosa et al. [3, 6]. That is, although the prognosis is better than pancreatic ductal adenocarcinoma, the majority of patients with acinar cell carcinoma will still succumb to disease. Currently, the only effective treatment is surgery for early stage resectable tumours, with the addition of platinum-based chemotherapy for the up to 20% of acinar cell carcinomas, which may be associated with somatic or germline BRCA mutations [7]. Therefore new therapies for patients with metastatic disease are required.
Recently large scale genomic projects have demonstrated that the molecular characteristics of acinar carcinoma are very different to pancreatic ductal adenocarcinoma [8,9,10]. In contrast to pancreatic ductal adenocarcinoma which very frequently harbours mutations in KRAS, TP53, CDKN2A, and SMAD4, somatic mutations in acinar carcinomas are very heterogeneous with recurrent mutation of individual genes being rare. However pathogenic gene rearrangements, rare in pancreatic ductal adenocarcinoma, appear to be relatively common in acinar cell carcinoma. For example, several studies have demonstrated gene rearrangements in BRAF in ~20% of acinar carcinomas [11,12,13,14]. Importantly these fusions, which cause downstream activation of the MAPK signalling pathway, are potentially targetable with currently available MEK inhibitors and therapeutic MEK inhibition (an established approach in melanoma) is under active investigation in BRAF gene rearranged acinar carcinomas [15].
The RET proto-oncogene located at chromosome 10q11.21 encodes for a ligand dependent receptor tyrosine kinase. Activating RET gene abnormalities are well recognized drivers of certain malignancies, with germline or somatic mutations being associated with up to 65% of medullary thyroid carcinomas and somatic fusions being found in 10–20% of papillary thyroid carcinomas [16, 17]. Apart from these tumours with a high incidence of RET gene abnormalities, RET mutations and fusions have been reported at a low incidence in a range of malignancies including 1–3% of non-small cell lung carcinomas and less than 1% of pancreatic carcinomas [16].
For some time mulitkinase inhibitors originally designed to target other tyrosine kinases but with non-selective action against RET including vandetinib, cabozantinib, lenvatinib, ponantinib, sunitinib, regorafenib and sorafenib have been available [16,17,18]. Results have been less impressive than with some other targeted therapies and tempered by side effects due to inhibition of other ‘off-target’ tyrosine kinases, however unequivocal responses have been reported particularly for cases associated with RET fusions [17, 19, 20].
Recently a new generation of highly specific small molecule RET tyrosine kinase inhibitors have been developed. These compounds which include LOXO-292 and BLU-667 have shown potential in early phase clinical trials [17, 21, 22]. For example in a recent and ongoing early phase basket trial for any malignancy harbouring RET fusions or mutations, LOXO-292 demonstrated an overall response rate of 77% in RET fusion positive malignancies of any lineage, with 92% of responses being ongoing and minimal toxicity [22, 23]. Of note two of the fusion positive patients in this trial were described as having ‘pancreatic cancer’ although the precise histology was not specified.
RET fusions are known to be rare in pancreatic malignancies. In one study, 1 of 160 (0.6%) of pancreatic cancers were shown to harbour a pathogenic RET fusion [16]. In the Australian Pancreatic Genome Initiative (APGI) and International Cancer Genome Consortium (ICGC) cohort of 456 pancreatic carcinomas only 1 (0.2%) RET fusion positive cancer was identified [24,25,26]. Of note we have reviewed the histology of this tumour and confirmed that this was an acinar cell carcinoma harbouring a CCDC6-RET fusion rather than a conventional pancreatic adenocarcinoma. Given the relatively low incidence of acinar carcinomas, the fact that it is molecularly distinct from pancreatic ductal adenocarcinomas and the finding that acinar cell carcinomas are known to be associated with a high incidence of BRAF fusions, we therefore postulated that pathogenic RET fusions would be relatively common in acinar cell carcinomas.
For these reasons we sought to investigate the incidence of RET gene rearrangements in a large cohort of acinar cell carcinomas and also took the opportunity to screen the same cohort for BRAF gene rearrangements.
Methods
We searched the institutional databases of multiple centres from Australia, one centre from Amsterdam, Netherlands and one centre from Bern, Switzerland for all cases recorded as pancreatic acinar cell carcinoma, pancreatoblastoma or mixed pancreatic acinar cell carcinoma-neurondocrine tumour. All cases were independently reviewed centrally by two surgical pathologists with expertize in pancreatic pathology to confirm the diagnosis (AJG and AC). Inclusion criteria included the expression of trypsin and BCL10 by immunohistochemistry and sufficient tumour in formalin-fixed paraffin-embedded tissue blocks for further testing. All tumours also underwent beta-catenin immunohistochemistry to highlight the squamous morule component of pancreatoblastomas.
Clinicopathological data collected included tumour size, age, sex, date of surgery, type of operation, type of sample, size of tumour, metastasis at presentation, lymph node involvement, AJCC pathological stage, and mitotic rate per 10 high power fields (2 mm2).
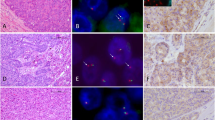
Fluorescent in-situ hybridization (FISH) was performed on formalin-fixed paraffin-embedded sections of tumours using Zytolight SPEC RET and BRAF dual colour break-apart probes (ZytoVision, Bremerhaven, Germany) according to the manufacturer’s instructions. Both probes include green and orange labelled probes that directly hybridize to the genes. The orange probe hybridizes to the proximal end and the green probe hybridizes to the distal end of the 10q11.21 and 7q34 chromosomal regions harbouring the RET and BRAF genes respectively. A rearrangement negative tumour cell was defined by the detection of fused green and orange signals. A rearrangement positive tumour cell was defined as the presence of a split green and orange signal, which was at least two signals diameter apart (Fig. 1b, f). At least 50 tumour nuclei were counted, and for a case to be considered as positive, at least 15% of the tumour nuclei were required to demonstrate split signals. The BRAF FISH status of 11 cases has previously been presented in a technical report [14].
a A RET-rearranged acinar cell carcinoma (H&E original magnification ×200). b RET FISH using break-apart probe shows the presence of one fused orange–green signal (black arrow) and one separated orange–green signal (white arrow) in the tumour cells, confirming RET rearrangement. This pattern was present in >15% of tumour cells. (original ×1000) c BCL10 IHC demonstrating positive cytoplasmic staining in acinar cell carcinoma (original magnification ×600). d Trypsin IHC demonstrating positive cytoplasmic staining in acinar cell carcinoma (original magnification ×600). e A BRAF-rearranged acinar cell carcinoma (H&E original ×200). f BRAF FISH using break-apart probe. The presence of a split signal (white arrow) confirms BRAF rearrangement in a tumour cell, which is usually accompanied by a fused signal (black arrow) (original ×1000)
RNA from formalin-fixed paraffin-embedded tumour samples was extracted using the Qiagen MiRNeasy FFPE kit using the manufacturer’s instructions. RNA library was prepared using a custom designed Ion AmpliSeq RNA panel (Thermo Fisher Scientific, Waltham, MA, USA) which covers transcripts for fusion partners with RET, CCDC6, PRKARIA, NCOA4, GOLGA5, TRIM24 and TRIM33, before sequencing on the Ion GeneStudio S5 sequencer (Thermofisher Scientific). Sequence data were then analysed using the Ion report software (Thermofisher Scientific). RNA library was also prepared using a hybridization capture-based Trusight® RNA fusion panel (Illumina, San Diego, CA, USA) before sequencing on the Illumina Miseq sequencer, and sequencing data were analysed using the Illumina Basespace RNA-Seq alignment App.
Statistical analysis was performed using IBM SPSS Statistics software v23. Continuous variables were compared using the Student’s t test and categorical variables were compared using the Fisher’s exact test. A p value of <0.05 was considered statistically significant.
Results
Of 40 cases of confirmed pancreatic acinar cell spectrum lesions with material available for FISH studies identified, 36 were pure acinar cell carcinomas, 3 were pancreatoblastomas and one case was a mixed acinar cell carcinoma-neuroendocrine tumour. Beta-catenin immunohistochemistry demonstrated aberrant positive nuclear staining only in the squamous morular component of the three pancreatoblastomas. No other cases demonstrated nuclear staining for beta-catenin. Thirty-six cases demonstrated diffuse positive staining for trypsin and BCL10, whilst four cases showed focal staining for both markers. The clinicopathological characteristics of the cohort are summarized in Table 1. Briefly, there were 30 males and ten females. The median age was 66 years (mean 61 years, range 17–86 years). The median tumour size was 37 mm (mean 45 mm, range 13–140 mm). Nine patients presented with stage I disease, 14 with stage II disease and 12 with stage IV disease. The median mitotic rate was 5 per 10 high power fields (2 mm2) (mean 11/hpf; range 1–44/10 hpf).
FISH testing for RET was successful in all 40 cases. BRAF testing failed in only one case where the signal intensity was too weak for interpretation. Rearrangement of the RET gene was found in three cases (7.5%) and BRAF rearrangement found in five cases (12.5%) (Fig. 1). Of note one of the three RET fusion positive cases (PACC12) was later confirmed to be the APGI case known to harbour a RET-CCDC6 fusion.
Gene rearrangements were only identified in pure acinar cell carcinomas and the presence of BRAF and RET rearrangements were mutually exclusive. Morphologically, cases with RET or BRAF rearrangement did not show any distinctive features compared to cases not harbouring gene rearrangements, and all rearranged cases demonstrated diffuse strong expression for BCL10 and trypsin. The clinicopathological characteristics of the eight rearranged cases are summarized in Table 2.
Since only pure acinar cell carcinomas were positive for RET or BRAF rearrangements, univariate analysis was performed comparing gene rearrangement positive and rearrangement negative cases in the 36 pure acinar cell carcinoma cases. There was no significant difference between the two groups based on sex (p = 0.558), age at presentation (p = 0.542), size (p = 0.638), metastasis at presentation (p = 0.827), nodal involvement at presentation (p = 0.064), pathological stage (p = 0.171) and mitotic rate (p = 0.332) (Table 3).
The three RET-rearranged acinar carcinomas detected by FISH were sequenced using a custom designed Ion AmpliSeq RNA panel (Thermo Fisher Scientific) which covers transcripts for fusion partners with CCDC6, PRKARIA, NCOA4, GOLGA5, TRIM24, TRIM33. Unfortunately PACC12 was inadequate for sequencing due to low RNA quantity and quality. The two other samples (PACC14 and PACC33) were negative for the RET fusion transcripts covered by the panel. As PACC12 was known to harbour a RET-CCDC6 fusion by the APGI data, only PACC14 and PACC33 were further examined using the Trusight RNA fusion panel. However due to poor coverage of exons, the results were inconclusive.
Discussion
This is the first study to systematically screen a large cohort of acinar cell carcinomas specifically for RET gene rearrangements. Given that the reported incidence of RET gene rearrangements in all pancreatic cancers is as low as 0.2 to 0.6% [16, 24,25,26], our novel finding of a relatively high incidence (3 of 36, 8%) in pure acinar cell carcinomas strongly supports our central hypothesis—that RET gene rearrangements are highly over-represented in acinar cell carcinomas. Our concurrent confirmation of an even higher incidence of BRAF gene rearrangements in pure acinar cell carcinomas (5 of 36, 14%) is in keeping with other studies that have reported an incidence of up to 20% and supports the established finding that BRAF fusions are also highly over-represented in acinar cell carcinomass [11, 12, 14].
We note that in our cohort, BRAF and RET gene rearrangements were mutually exclusive and only occurred in pure acinar carcinomas. This exclusiveness was also found in a pan-cancer cohort of solid tumours (n = 4871) where RET aberrations were also mutually exclusive with gene aberrations that affect MAPK signalling pathway such as (KRAS, NRAS, BRAF and NF1) [16]. Given the high frequency of KRAS mutations in conventional pancreatic adenocarcinomas (more than 93% in our centre) [24,25,26], this further supports our contention that RET gene rearrangements are particularly over-represented in acinar carcinomas and very uncommon in conventional pancreatic ductal adenocarcinomas. Indeed we think it is likely that at least some if not the majority of ‘pancreatic cancers’ (reported without further specification of histology) in studies of carcinomas of multiple primary sites and the early basket clinical trials of the novel small molecule RET inhibitor LOXO-292 may represent acinar carcinomas [16, 22, 23]. Of course only specific histologic review of the fusion positive cases from these studies would definitively prove this hypothesis. In a similar vein we acknowledge the recent publication by Singhi et al., where n = 4 (0.1%) of 3738 pancreatic adenocarcinomas undergoing molecular testing were found to have a RET fusion (all were KRAS wild type) [27] and note that in this study, although there was secondary pathological review based on H&E stained sections and scanned images, the authors did not state if immunohistochemistry for BCL10 or trypsin was performed to exclude the possibility that these cases may represent acinar carcinomas. This is important because we suspect that acinar cell carcinomas may be under-recognized. For example we previously reviewed an APGI/ICGC pancreatic carcinoma originally classified as pancreatic ductal adenocarcinoma after it was found to be KRAS wild type and harbour an oncogenic SDK1-BRAF fusion [24,25,26]. Upon review we demonstrated that it was actually a misclassified acinar cell carcinoma based on both morphology and diffuse strong expression of BCL10 and trypsin (data not shown). We therefore recommend that pathologists should have a low threshold for considering the diagnosis of acinar cell carcinoma and performing BCL10 and trypsin immunohistochemistry.
There are different methods for detecting gene rearrangements in malignancies including whole-genome and transcriptome sequencing, RNA sequencing, real-time polymerase chain reaction (RT-PCR) and FISH. Each of these techniques have their advantages and disadvantages in clinical practice. FISH is appealing in the diagnostic surgical pathology laboratory as it can be readily performed on formalin-fixed paraffin-embedded tissue (including archived specimens) and has the advantage of low cost, minimal tissue requirement, rapid turnaround and, importantly, a low failure rate. Indeed, we were able to detect the presence or absence of BRAF or RET gene rearrangement using FISH break-apart probes in all cases except one due to poor signal quality (1/80), yet had significant difficulties in achieving results for fusion testing using the RNA fusion panels. However one very significant disadvantage of FISH is that it detects only the presence of a gene rearrangement. Whilst it is assumed that this usually reflects an oncogenic fusion gene, FISH studies do not definitively prove a fusion or identify the partner gene. At the time of writing 12 different RET fusions partners have been identified (TRIM33, NCOA4, KIF5B, CCDC6, PRKARIA, GOLGA5, TRIM24, KTN1, RAB61P2, MBD1, RFP, SQSTM1) but there are also reports of gene rearrangements without identified partners [16, 28]. Our amplicon-based gene panel included only six of these genes (CCDC6, PRKARIA, NCOA4, GOLGA5, TRIM24, TRIM33), therefore it is likely that the RET fusions in our samples were not covered by this panel and hence not detected. Unfortunately using a comprehensive 507 gene hybridization capture-based panel we could not obtain adequate regional coverage of RET to adequately interpret the result. Therefore whilst we were confident with the identification of a fusion partner for one case (RET-CCDC6 in case PACC12), we could not identify a fusion partner for the other RET gene rearranged cases. This is important because it is possible that these FISH aberrations may reflect genomic instability at the chromosomal locus rather than a true driving fusion gene. Furthermore current entry criteria for many clinical trials (including the LOXO trial) requires identification of a gene fusion, with FISH studies alone being considered insufficient. Therefore, whilst FISH testing has some advantages, this approach is not without its limitations compared to RET fusion analysis.
In conclusion, RET and BRAF rearrangements are found in ~20% of acinar cell carcinomas and can be readily identified in the routine clinical setting using FISH. The identification of RET gene aberrations is clinically highly significant, as they are already targetable using FDA-approved multikinase inhibitors and the subject of ongoing but promising clinical trials of more specific inhibitors. In the era of personalized medicine pathologists are often encouraged to perform pan-cancer panel testing for numerous molecular abnormalities regardless of morphology or histogenesis. We propose that basic morphology, with simple confirmatory immunohistochemistry for BCL10 and trypsin, can be used to diagnose acinar cell carcinoma and serve as a cost effective method to triage molecular testing for RET and BRAF gene rearrangements in pancreatic malignancies. We therefore recommend a low threshold for considering the diagnosis of acinar cell carcinoma and, in confirmed cases of acinar cell carcinoma, for testing for these gene rearrangements. At this stage this would be primarily to recruit patients for clinical trials of new targeted therapies. However given the provisional results of these clinical trials, probably in the near future RET and BRAF fusion testing and targeting will be part of routine clinical care for all acinar cell carcinomas.
References
Schmidt CM, Matos JM, Bentrem DJ, Talamonti MS, Lillemoe KD, Bilimoria KY. Acinar cell carcinoma of the pancreas in the United States: prognostic factors and comparison to ductal adenocarcinoma. J Gastrointest Surg. 2008;12:2078–86.
Klimstra DS, Hruban RH, Klöppel G, Morohoshi T, Ohike N Acinar cell neoplasms of the pancreas. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO classification of tumours of the digestive system. 4th ed. Lyon: IARC Press; 2010. p. 314–8.
Klimstra DS, Adsay V. Acinar neoplasms of the pancreas—a summary of 25 years of research. Semin Diagn Pathol. 2016;33:307–18.
Wisnoski NC, Townsend CM Jr., Nealon WH, Freeman JL, Riall TS. 672 patients with acinar cell carcinoma of the pancreas: a population-based comparison to pancreatic adenocarcinoma. Surgery. 2008;144:141–8.
Klimstra DS, Heffess CS, Oertel JE, Rosai J. Acinar cell carcinoma of the pancreas. A clinicopathologic study of 28 cases. Am J Surg Pathol. 1992;16:815–37.
La Rosa S, Adsay V, Albarello L, Asioli S, Casnedi S, Franzi F, et al. Clinicopathologic study of 62 acinar cell carcinomas of the pancreas: insights into the morphology and immunophenotype and search for prognostic markers. Am J Surg Pathol. 2012;36:1782–95.
Kryklyva V, Haj Mohammad N, Morsink FHM, Ligtenberg MJL, Offerhaus GJA, Nagtegaal ID, et al. Pancreatic acinar cell carcinoma is associated with BRCA2 germline mutations: a case report and literature review. Cancer Biol Ther. 2019;20:949–55.
Furukawa T, Sakamoto H, Takeuchi S, Ameri M, Kuboki Y, Yamamoto T, et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci Rep. 2015;5:8829.
Jakel C, Bergmann F, Toth R, Assenov Y, van der Duin D, Strobel O, et al. Genome-wide genetic and epigenetic analyses of pancreatic acinar cell carcinomas reveal aberrations in genome stability. Nat Commun. 2017;8:1323.
Bergmann F, Aulmann S, Sipos B, Kloor M, von Heydebreck A, Schweipert J, et al. Acinar cell carcinomas of the pancreas: a molecular analysis in a series of 57 cases. Virchows Arch. 2014;465:661–72.
Chmielecki J, Hutchinson KE, Frampton GM, Chalmers ZR, Johnson A, Shi C, et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov. 2014;4:1398–405.
Wang L, Basturk O, Wang J, Benayed R, Middha S, Zehir A, et al. A FISH assay efficiently screens for BRAF gene rearrangements in pancreatic acinar-type neoplasms. Mod Pathol. 2018;31:132–40.
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138:881–90.
Chou A, Kim Y, Samra JS, Pajic M, Gill AJ. BRAF gene rearrangements can be identified by FISH studies in pancreatic acinar cell carcinoma. Pathology. 2018;50:345–8.
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846.
Kato S, Subbiah V, Marchlik E, Elkin SK, Carter JL, Kurzrock R. RET aberrations in diverse cancers: next-generation sequencing of 4,871 patients. Clin Cancer Res. 2017;23:1988–97.
Subbiah V, Velcheti V, Tuch BB, Ebata K, Busaidy NL, Cabanillas ME, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol. 2018;29:1869–76.
Roskoski R Jr., Sadeghi-Nejad A. Role of RET protein-tyrosine kinase inhibitors in the treatment RET-driven thyroid and lung cancers. Pharm Res. 2018;128:1–17.
Yoh K, Seto T, Satouchi M, Nishio M, Yamamoto N, Murakami H, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet. Respir Med. 2017;5:42–50.
Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol. 2016;17:1653–60.
Subbiah V, Gainor JF, Rahal R, Brubaker JD, Kim JL, Maynard M, et al. Precision targeted therapy with BLU-667 for RET-driven cancers. Cancer Discov. 2018;8:836–49.
Drilon AESV, Oxnard GR, et al. A Phase 1 study of LOXO-292, a potent and highly selective RET inhibitor, in patients with RET-altered cancers. J Clin Oncol. 2018;36:102–10.
Dolgin E. LOXO-292 Reins in RET-driven tumors. Cancer Discov. 2018;8:904–5.
Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405.
Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501.
Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Singhi AD, George B, Greenbowe JR, Chung J, Suh J, Maitra A, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156:2242–53 e4.
Santoro M, Melillo RM, Fusco A. RET/PTC activation in papillary thyroid carcinoma: European Journal of Endocrinology Prize Lecture. Eur J Endocrinol. 2006;155:645–53.
Acknowledgements
AC is supported by the CINSW ECF (AC0430). This study was supported by the Avner Australian Pancreatic Cancer Genome Initiative (APGI) BioResource partially funded by the Avner Pancreatic Cancer Foundation Grant, www.avnersfoundation.org.au.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chou, A., Brown, I.S., Kumarasinghe, M.P. et al. RET gene rearrangements occur in a subset of pancreatic acinar cell carcinomas. Mod Pathol 33, 657–664 (2020). https://doi.org/10.1038/s41379-019-0373-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0373-y
This article is cited by
-
Post-therapy emergence of an NBN reversion mutation in a patient with pancreatic acinar cell carcinoma
npj Precision Oncology (2024)
-
Pancreatoblastomas and mixed and pure acinar cell carcinomas share epigenetic signatures distinct from other neoplasms of the pancreas
Modern Pathology (2022)
-
RAF1 rearrangements are common in pancreatic acinar cell carcinomas
Modern Pathology (2020)