Abstract
Superficial/cutaneous malignant peripheral nerve sheath tumor is a rare soft tissue neoplasm that shares morphological, immunohistochemical, and molecular features with spindle/desmoplastic melanoma. We aimed to identify a methylome signature to distinguish these two entities. We analyzed 15 cases of spindle/desmoplastic melanoma and 15 cases of cutaneous malignant peripheral nerve sheath tumor in 23 men and 7 women. DNA from formalin-fixed, paraffin-embedded tissues was extracted and processed using the Illumina Infinium Methylation EPIC array interrogating 866,562 CpG sites. Using a home-grown informatics pipeline, we identified differentially methylated positions between the two entities. Functional network analysis for enrichment signatures was performed using DAVID tools. Identified differentially methylated positions were compared with the Cancer Genome Atlas’s cutaneous melanoma dataset and a recently published malignant peripheral nerve sheath tumor dataset to assess the specificity of the identified signature. Unsupervised hierarchical clustering showed different patterns of methylation in cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma. Two probes, cg20783223 and cg13332552, colocalized in the promoter region of BCAT1 and miR-2504. Pathway analysis highlighted enrichment in a subset of genes involved in breast and gastric cancer centered on BCAT1 and downstream activated genes in the mTOR pathway. Our study identifies BCAT1 as a novel methylome signature distinguishing spindle/desmoplastic melanoma from cutaneous malignant peripheral nerve sheath tumor.
Similar content being viewed by others
Introduction
Cutaneous and subcutaneous spindle cell neoplasms cause diagnostic challenges even for the most experienced dermatopathologists. Within this group of tumors, superficial (supra-fascial) cutaneous and subcutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma are among the most challenging to diagnose.
Cutaneous malignant peripheral nerve sheath tumor is a variant of malignant peripheral nerve sheath tumor, a rare, aggressive, heterogeneous soft tissue neoplasm accounting for 4% of all sarcomas [1]. Malignant peripheral nerve sheath tumor has a propensity to arise in the deep soft tissue of the lower and upper extremities and trunk. Up to 50% of malignant peripheral nerve sheath tumor cases arise in patients with neurofibromatosis type 1; the remaining cases occur sporadically or in patients with previous radiation therapy [1]. Cutaneous malignant peripheral nerve sheath tumor is an extremely rare neoplasm arising mostly in association with neurofibroma [2]. Some reports showed indolent behavior of cutaneous malignant peripheral nerve sheath tumor, but larger studies showed that cutaneous malignant peripheral nerve sheath tumor exhibits a propensity for local recurrence and metastasis akin to that of deep malignant peripheral nerve sheath tumor [3]. Morphologically, low- to intermediate-grade cutaneous malignant peripheral nerve sheath tumor exhibits a spindle cellular proliferation with nuclear crowding and relatively moderate atypia [2]. High-grade cutaneous malignant peripheral nerve sheath tumor exhibits alternating hypocellular and hypercellular fascicles of spindle cells with areas of geographic necrosis [2].
Spindle/desmoplastic melanoma is a rare variant of melanoma affecting sun-damaged skin and characterized by a propensity for local recurrence and local lymph node metastasis [4]. Morphologically, spindle/desmoplastic melanoma encompasses a spectrum ranging from spindle/mixed desmoplastic melanoma, in which the tumor shows fascicles of spindle cells in a sheet-like distribution with minimal stroma, to pure desmoplastic melanoma, in which the collagenous stroma occupies more than 90% of the tumor surface [5, 6]. Pure desmoplastic melanoma rarely disseminates to regional lymph nodes and is associated with a better outcome than spindle/mixed desmoplastic melanoma [6].
Recent advances in molecular technologies have enabled investigators to explore the molecular landscape of cutaneous malignant peripheral nerve sheath tumor and melanomas. However, published studies focusing on differentiating between cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma are scarce. In a recent study, differential gene expression analysis showed that both cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma exhibited significantly perturbed immune pathway components, including nuclear factor-κB, JAK-STAT, and CXCL12-CXCR4, and overexpression of both CD274 and CTLA4 [7]. However, although this previous study identified important molecular and immune pathways that are targetable for patients with unresectable tumors, it did not identify a distinct molecular signature that could be used to accurately distinguish between spindle/desmoplastic melanoma and cutaneous malignant peripheral nerve sheath tumor.
In the study reported herein, we used a genomewide methylation array including probes targeting 866,562 CpG islands in an attempt to identify a methylome signature that could distinguish cutaneous malignant peripheral nerve sheath tumor from spindle/desmoplastic melanoma. In the first phase of this work, we identified a novel methylome signature distinguishing cutaneous malignant peripheral nerve sheath tumor from spindle/desmoplastic melanoma. In the second phase, we compared our results with the methylation profiles of melanomas in the Cancer Genome Atlas cohort (467 samples) [8] and deep malignant peripheral nerve sheath tumors in a German cohort (27 samples) [9].
Materials and methods
Case selection
After institutional review board approval at the 2 collaborating institutions (University of Texas MD Anderson Cancer Center and NYU Langone Medical Center), 15 cases of cutaneous malignant peripheral nerve sheath tumor and 15 cases of spindle/desmoplastic melanoma were selected from among patients diagnosed during the period from 2000 through 2015. All available slides for each case were reviewed by two pathologists (GJ and VGP) certified in dermatopathology, and bone and soft tissue pathology, to confirm the diagnosis and select the most viable tumor areas for DNA extraction. All spindle/desmoplastic melanoma cases were from resected tumors with an intra-epidermal in situ component; S100 (diffuse and strong nuclear and cytoplasmic), nestin, and SOX10 positivity (all staining performed as part of the initial clinical workup); and moderate-to-severe solar elastosis. Criteria for diagnosing cutaneous malignant peripheral nerve sheath tumor included the following: (1) lack of intra-epidermal in situ component and only dermal/subcutaneous involvement; (2) lack of moderate-to-severe solar elastosis; (3) patchy weak-to-moderate nuclear and cytoplasmic S100 positivity; and (4) the presence of a remnant neurofibroma from which the tumor arose (whenever applicable). In addition, any lesion exhibiting severe cytologic atypia with geographic necrosis and brisk mitotic activity was considered a cutaneous malignant peripheral nerve sheath tumor. The study was conducted at The University of Texas MD Anderson Cancer Center and approved by the Institutional review Board in accordance with all local and federal regulations.
Tissue selection, nucleic acid isolation, and DNA processing
For each specimen, ten 10 μm-thick formalin-fixed, paraffin-embedded sections were cut from a single representative block. Macrodissection was performed with a scalpel and focused on the areas of greatest tumor cell density to try to ensure that tumor cells accounted for at least 70% of the material analyzed for each case. DNA was recovered using the PicoPure DNA extraction kit (Thermo Fisher, Life Technologies), which enables recovery of genomic DNA from formalin-fixed, paraffin-embedded tissue. DNA was then subjected to bead purification with the Sphere quality control kit (Thermo Fisher, Life Technologies). DNA was quantified using the Qubit 2.0 fluorometer (Life Technologies). The DNA was bisulfite converted using the EZ-96 DNA methylation kit (Zymo Research, Irvine, CA). Extracted DNA was restored using the Infinium HD formalin-fixed, paraffin-embedded DNA restore kit (Illumina, San Diego, CA) prior to hybridization on the bead chips provided by the manufacturer (Illumina).
Differential methylation analysis and unsupervised hierarchical clustering
The Infinium Methylation EPIC array (Illumina) was used to determine the DNA methylation status of 866,562 CpG sites, following the manufacturer’s instructions. Minfi R package was used to process and analyze the methylation data [10]. Using minfi, the probes were quantile normalized and background adjusted. The resulting set of samples and probes was used for differentially methylated probes analysis. The differentially methylated probes finder function in minfi was used to identify differentially methylated probes for three different comparisons: (1) across all groups of cell populations, (2) spindle/desmoplastic melanoma vs. malignant peripheral nerve sheath tumor, and (3) malignant peripheral nerve sheath tumor vs. spindle/desmoplastic melanoma. Probes with statistical significance using Benjamini–Hochberg false discovery rate q cutoff of q < 0.05 were considered most significant, and corresponding heatmaps are shown. β-Values for all 866,562 CpG sites tested were defined as the ratio of fluorescence intensity of the methylated probe over the overall intensity of probes. β < 0.2 indicated hypomethylation (blue); β > 0.8 indicated hypermethylation (red). All statistical analysis and modeling was performed using the open-source software R (https://www.r-project.org/). All graphs and heatmaps were generated using the R package. Unsupervised hierarchical clustering was done with Euclidean measure for distance matrix and complete agglomeration method for clustering was used for unsupervised hierarchical clustering.
Functional genomic pathway analysis
All probes included in the array were annotated using the HumanMethylation850 manifest provided by the manufacturer (MethylationEPIC_v-1-0_B4; Illumina). Genomic information, including DNA sequence and coordinates of gene-coding regions, was obtained from the University of California Santa Cruz Genome Browser database [11]. All probes covering promoters and enhancers of coding genes were filtered in and considered for the enrichment pathway network analysis. This rationale was adopted to limit nonspecific enrichment pathway results that may occur when all coding and noncoding genes are included. Using the R package, we ran in parallel Cluster profiler and the DAVID Bioinformatics Resources interrogating 5867 genes in order to detect differentially methylated genes [12,13,14]. The enrichment test is based on the Fisher exact test, which indicates if the overlap between genes in a cluster and in a Gene Ontology term is higher than expected by chance. All sources of interaction evidence are calibrated against previous knowledge using the high-level functional groupings provided by the manually curated Kyoto Encyclopedia of Genes and Genomes pathway map [15].
Comparison with the Cancer Genome Atlas cutaneous melanoma data and deep malignant peripheral nerve sheath tumor data
To test the specificity of the methylome signature, we compared our top 100, 500, 1000, and 5000 desmoplastic melanoma probes with those obtained from the clustering analysis of the Cancer Genome Atlas methylation data on melanoma and recently published methylation data on 27 deep malignant peripheral nerve sheath tumors (generated using the HumanMethylation450). Raw signal intensity data (IDAT) files from the Cancer Genome Atlas (n = 470) were downloaded from the legacy archives of the Genomic Data Commons [16]. Similarly, IDAT files were obtained from Röhrich et al. [9] (Institute of Pathology, University of Heidelberg, Heidelberg, Germany). We developed an algorithm to extract the common probes common to both HumanMethylation450 and EPIC HD850 for the comparative analysis.
Data access
IDAT files of our tumor set (30 samples) have been deposited in Gene Expression Omnibus, accession number GSE112308. The R script used for the analysis has been deposited on Github (https://github.com/varshivasu7/minfi_Illumina_methylation).
Results
Clinical and pathological findings
Clinical and pathological findings for the patients with cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma are summarized in Table 1, and photomicrographs of a case of spindle/desmoplastic melanoma and cutaneous malignant peripheral nerve sheath tumor case 12, which clustered with the spindle/desmoplastic melanoma group, are shown in Fig. 1. Twelve of the 15 patients (80%) with cutaneous malignant peripheral nerve sheath tumor and 11 of the 15 patients (73%) with spindle/desmoplastic melanoma were men. Nine of the 15 (60%) cutaneous malignant peripheral nerve sheath tumors arose in dermal and subcutaneous plexiform and/or diffuse neurofibromas. Diffuse immunoreactivity with S100 and SOX10 was noted in all spindle/desmoplastic melanomas and background neurofibromas. In contrast, only patchy weak-to-moderate immunoreactivity with S100 was noted in the cutaneous malignant peripheral nerve sheath tumors.
Photomicrographs of a selected case of spindle/desmoplastic melanoma (a, c) and cutaneous malignant peripheral nerve sheath tumor case 12 (b, d), which clustered with the spindle/desmoplastic melanoma group. (a, b) Hematoxylin–eosin staining, × 200 magnification, showing morphological features. Both tumors show spindle cell morphology with prominent cytologic atypia. (c) Spindle/desmoplastic melanoma with nuclear atypia and perineural invasion (star) (hematoxylin–eosin staining, × 200 magnification). (d) High-grade cutaneous malignant peripheral nerve sheath tumor with mitotic figures (arrow) (hematoxylin–eosin staining, × 400 magnification)
Differential methylation of CpG sites related to coding and noncoding genes
We identified 91,085 probes, including 66,389 promoter-associated probes, that were significantly differentially methylated (p < 0.05, q < 0.01) between cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma (Supplementary Table S1). Subsequent analysis focusing on protein-coding genes within the 66,389 promoter-associated probes identified 699 probes that were significantly differentially methylated between cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma (Supplementary Table S2).
Distinct methylation profiles in cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma revealed by unsupervised hierarchical clustering
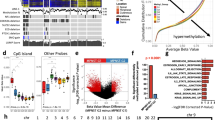
We selected the top 5000, 1000, 500, and 100 differentially methylated probes corresponding to probes with greatest F-test value and lowest associated p- and q-values (Supplementary Table S1). Given the high correlations among all four sets of probes (R2 = 0.98), we present the heatmap based on the top 100 differentially methylated probe sets (Fig. 2). Unsupervised hierarchical clustering focusing on the top 100 differentially methylated probe sets showed different patterns of methylation in cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma. Both patterns of methylation in these tumors were distinct from the patterns of methylation in the Cancer Genome Atlas cutaneous melanoma cohort and the deep malignant peripheral nerve sheath tumor cohort obtained from Röhrich et al. [9]. Probes that were methylated in cutaneous malignant peripheral nerve sheath tumor were unmethylated in spindle/desmoplastic melanoma and vice versa (Fig. 2a). Two probes colocalized (covered regions) in genes of interest: probe cg20783223 and probe cg13332552 (see Discussion below).
(a) Unsupervised hierarchical clustering heatmap with different β-scores for the 100 most significantly differentially methylated gene-coding and non-gene-coding CpG islands [i.e., lowest p- and q-values (p < 0.05; q < 0.01)]. The heatmap shows clear segregation of spindle/desmoplastic melanoma and cutaneous malignant peripheral nerve sheath tumor. Hypermethylated loci in one tumor are hypomethylated in the other and vice versa. Two probes, cg13332552 and cg20783223, indicated by the upper and lower arrows, respectively, indicate the miR-2504 promoter site and the BCAT1 promoter site. The data from this heatmap can be used in challenging clinical cases to classify ambiguous superficial spindle cell tumors. (b) Two-dimensional principal component analysis showing the distribution of cutaneous malignant peripheral nerve sheath tumor cases (orange dots) and spindle/desmoplastic melanoma cases (green dots) classified on the basis of differentially methylated probes adjusted for age and sex. The two cutaneous malignant peripheral nerve sheath tumor cases (#12 and #13) that were incorrectly classified in the spindle/desmoplastic melanoma group rather than in the cutaneous malignant peripheral nerve sheath tumor group are indicated with black arrows
Performance of the methylation signature in distinguishing spindle/desmoplastic melanoma from cutaneous malignant peripheral nerve sheath tumor
When this methylome signature was applied using the top 100, 500, and 1000 differentially methylated probes as a diagnostic tool to distinguish between spindle/desmoplastic melanoma and cutaneous malignant peripheral nerve sheath tumor, 13 of 15 cutaneous malignant peripheral nerve sheath tumors and 15 of 15 spindle/desmoplastic melanomas clustered in their respective regions, yielding sensitivity of 86.6% in the detection of cutaneous malignant peripheral nerve sheath tumor and 100% in the detection of spindle/desmoplastic melanomas (Fig. 2b).
Functional pathway analysis
Pathway enrichment analysis identified two gene sets that enriched and overlapped with our differentially methylated probe set for protein-coding genes: 3968 genes known to be enriched in breast cancer and 7318 genes known to be enriched in gastric cancer (Fig. 3).
Functional genomic pathway analysis. Summary of pathway network analysis highlights the relationship between gene sets enriched in cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma and other oncologic processes. Note the very significant enrichment for gastric cancer-related and breast cancer-related gene sets. These findings are in line with the differentially methylated probes results, which highlighted BCAT1 as a protein-coding gene that regulates the downstream mTOR and WNT pathways in both cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma. Y axis, the different gene sets with significant overlap with the probe sets/genes. X axis, the ratio of the number of studied genes to the total number of genes included in the particular gene set (Supplementary Table 2). The dot sizes are proportional to the number of overlapping genes. The dot colors show the adjusted p-value for false discovery rate ranging from 0.005 to 0.025 (p < 0.05)
Discussion
Probe cg20783223 is localized on chromosome 12 in the promoter region of BCAT1 approximately 200 bp from the transcription start site. Probes in close proximity to the transcriptional start (200 bp) site generally correlate strongly with variations in gene expression, whereas probes further downstream of the transcription start site (i.e., gene body and 3′-untranslated region (UTR)) do not [17]. The BCAT1 promoter exhibited significant high methylation in the spindle/desmoplastic melanoma group but was hypomethylated in the cutaneous malignant peripheral nerve sheath tumor group. BCAT1 promoter hypomethylation enhances gene expression and subsequent protein expression; this was shown in previous studies in epithelial ovarian carcinomas [18]. Furthermore, BCAT1 suppression led to significantly prolonged survival time in experimental models of advanced peritoneal ovarian carcinoma and breast cancer cells, highlighting its role in oncogenesis [18, 19]. BCAT1 (branched-chain aminotransferase 1) is a pivotal enzyme in the leucine catabolic pathway that promotes tumor growth through activation of the mammalian target of rapamycin (mTOR) pathway [19]. Recent studies showed that BCAT1 can be engineered to induce leucine deprivation in the tumor microenvironment, yielding an anti-cancer effect [20]. The finding of BCAT1 hypomethylation in cutaneous malignant peripheral nerve sheath tumor and the more aggressive course identified in this patient cohort (75% of patients died of disease) in comparison with the spindle/desmoplastic melanoma group (only 6% of patients died of disease) further highlights the role of BCAT1 in tumor progression and aggressiveness. In an ongoing study, we are investigating BCAT1 expression as a potential immunohistochemical biomarker for cutaneous malignant peripheral nerve sheath tumor and as a marker differentiating between cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma.
Probe cg13332552 is localized on chromosome 4 in the 5′-UTR region of miR-2504. This probe exhibited high methylation in the cutaneous malignant peripheral nerve sheath tumor group and hypomethylation in the spindle/desmoplastic melanoma group. This finding is in line with the known function of miR-2504, which showed maximal impact on cell invasion in experimental cell line invasion studies [21]. We further investigated the difference in the methylation profile based on the β-score (methylation score) between spindle/desmoplastic melanoma and pure desmoplastic melanoma, and found no statistical difference (p = 0.44). This further corroborates reported results from our previous study on the same cohort of spindle/desmoplastic melanoma and pure desmoplastic melanomas that showed similar gene expression signature for the tumor microenvironment with an enrichment for both JAK/STAT and nuclear factor-κB pathways and similar molecular events characterized by the high prevalence of TSC1/2 deleterious events, which were detected in up to 53% of the analyzed cases [7]. These findings suggest that while spindle/desmoplastic melanoma and pure desmoplastic melanoma show different morphology and clinical outcomes, they share similar molecular and epigenetic events.
The finding of two outlier cutaneous malignant peripheral nerve sheath tumor cases that clustered with the spindle/desmoplastic melanoma group is intriguing. Both of these tumors were confirmed histologically to be high-grade cutaneous malignant peripheral nerve sheath tumors, and both occurred in patients with neurofibromatosis type 1 in a background of atypical neurofibromas. Both tumors exhibited variable patchy staining for S100 and were positive for SOX10. Both patients had an aggressive disease course; one had a lung metastasis detected at 18 months, and the other (with specimens shown in Fig. 1b, d) died of cutaneous malignant peripheral nerve sheath tumor at 5 months. The aggressive course of cutaneous malignant peripheral nerve sheath tumors in those two patients (although the tumors were classified under “spindle/desmoplastic melanoma methylation class”) highlights the epigenetic heterogeneity in cutaneous malignant peripheral nerve sheath tumors, a phenomenon previously described in Ewing sarcomas [22]. Indeed, DNA methylation differences between cutaneous malignant peripheral nerve sheath tumors show a continuous spectrum of mesenchymal cell signatures that extends from spindle/desmoplastic melanoma to cutaneous malignant peripheral nerve sheath tumors. This would explain the mis-classification of these two tumors in the principal component analysis plot. Additional larger cohort studies should address overcoming methylation heterogeneity. None of the top 100, 500, 1000, or 5000 probes corresponding to differentially methylated probes in our two studied tumor groups overlapped with differentially methylated probes obtained from the comparative analysis with the Cancer Genome Atlas cutaneous melanoma cohort or deep malignant peripheral nerve sheath tumor cohort, indicating that the specificity of our signature was 100% (Supplementary Figures 1 and 2).
Among those genes, PIK3CA, from the mTOR pathway, and TCF7L1, WNT1, and AXIN2, from the WNT pathway, were among the most enriched (Supplementary Table 2). Interestingly, BCAT1 has a pivotal role in both pathways. Studies have shown that BCAT1 expression is a prognostic biomarker for gastric carcinomas, activates mTOR signaling, and facilitates growth and colony formation in breast cancer cells [19, 23]. Also, BCAT1 has an important regulatory role in the WNT pathway in colorectal carcinomas [24] (Supplementary Table 2).
In conclusion, we demonstrated a unique genomewide methylome signature enabling accurate differentiation between cutaneous malignant peripheral nerve sheath tumor and spindle/desmoplastic melanoma. These two entities cause diagnostic challenges even for the most experienced dermatopathologists. This comprehensive, rapid, cost-effective methylation array with coverage of 866,562 CpG sites provides unprecedented resolution assisting in differentiating between the two entities. Use of the methylation array is a robust approach resistant to factors that are known to interfere with gene expression analysis, including formalin fixation, ischemia time, and other factors. Further validation of this newly discovered methylome signature through analysis of larger cohorts is warranted.
References
Toro JR, Travis LB, Wu HJ, et al. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the Surveillance, Epidemiology and End Results program, 1978-2001: an analysis of 26,758 cases. Int J Cancer. 2006;119:2922–30.
Rodriguez FJ, Folpe AL, Giannini C, et al. Pathology of peripheral nerve sheath tumors: diagnostic overview and update on selected diagnostic problems. Acta Neuropathol. 2012;123:295–319.
Allison KH, Patel RM, Goldblum JR, et al. Superficial malignant peripheral nerve sheath tumor: a rare and challenging diagnosis. Am J Clin Pathol. 2005;124:685–92.
Chen LL, Jaimes N, Barker CA, et al. Desmoplastic melanoma: a review. J Am Acad Dermatol. 2013;68:825–33.
Busam KJ, Mujumdar U, Hummer AJ, et al. Cutaneous desmoplastic melanoma: reappraisal of morphologic heterogeneity and prognostic factors. Am J Surg Pathol. 2004;28:1518–25.
Hawkins WG, Busam KJ, Ben-Porat L, et al. Desmoplastic melanoma: a pathologically and clinically distinct form of cutaneous melanoma. Ann Surg Oncol. 2005;12:207–13.
Jour G, Andeen NK, Al-Rohil R, et al. Novel enriched pathways in superficial malignant peripheral nerve sheath tumours and spindle/desmoplastic melanomas. J Pathol. 2018;244:97–106.
NIH Genomic Data Commons Data Portal. https://portal.gdc.cancer.gov. 2017. Accessed 08/09/2018.
Rohrich M, Koelsche C, Schrimpf D, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131:877–87.
Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9.
Raney BJ, Dreszer TR, Barber GP, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–5.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–5.
Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20.
Martino D, Saffery R. Characteristics of DNA methylation and gene expression in regulatory features on the Infinium 450k Beadchip. BioRxiv. 2015;4:1–7. https://doi.org/10.1101/032862
Wang ZQ, Faddaoui A, Bachvarova M, et al. BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism. Oncotarget. 2015;6:31522–43.
Zhang L, Han J. Branched-chain amino acid transaminase 1 (BCAT1) promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Biochem Biophys Res Commun. 2017;486:224–31.
Sheen JH, Zoncu R, Kim D, et al. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–28.
Deng B, Wang B, Fang J, et al. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep. 2016;6:28301.
Sheffield NC, Pierron G, Klughammer J, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23:386–95.
Xu Y, Yu W, Yang T, et al. Overexpression of BCAT1 is a prognostic marker in gastric cancer. Hum Pathol. 2018;75:41–46.
Xie F, Xiang X, Huang Q, et al. Reciprocal control of lncRNA-BCAT1 and beta-catenin pathway reveals lncRNA-BCAT1 long non-coding RNA acts as a tumor suppressor in colorectal cancer. Oncotarget. 2017;8:23628–37.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jour, G., Vasudevaraja, V., Prieto, V.G. et al. BCAT1 and miR-2504: novel methylome signature distinguishes spindle/desmoplastic melanoma from superficial malignant peripheral nerve sheath tumor. Mod Pathol 32, 338–345 (2019). https://doi.org/10.1038/s41379-018-0146-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0146-z