Abstract
The breast stromal microenvironment is a pivotal factor in breast cancer development, growth and metastases. Although pathologists often detect morphologic changes in stroma by light microscopy, visual classification of such changes is subjective and non-quantitative, limiting its diagnostic utility. To gain insights into stromal changes associated with breast cancer, we applied automated machine learning techniques to digital images of 2387 hematoxylin and eosin stained tissue sections of benign and malignant image-guided breast biopsies performed to investigate mammographic abnormalities among 882 patients, ages 40–65 years, that were enrolled in the Breast Radiology Evaluation and Study of Tissues (BREAST) Stamp Project. Using deep convolutional neural networks, we trained an algorithm to discriminate between stroma surrounding invasive cancer and stroma from benign biopsies. In test sets (928 whole-slide images from 330 patients), this algorithm could distinguish biopsies diagnosed as invasive cancer from benign biopsies solely based on the stromal characteristics (area under the receiver operator characteristics curve = 0.962). Furthermore, without being trained specifically using ductal carcinoma in situ as an outcome, the algorithm detected tumor-associated stroma in greater amounts and at larger distances from grade 3 versus grade 1 ductal carcinoma in situ. Collectively, these results suggest that algorithms based on deep convolutional neural networks that evaluate only stroma may prove useful to classify breast biopsies and aid in understanding and evaluating the biology of breast lesions.
Similar content being viewed by others
Introduction
The diagnostic classification of benign breast diseases, putative breast cancer precursors, and breast cancer is based largely on the histopathological appearance and molecular characteristics of epithelial cells [1]. Although the appearance of breast stroma contributes to pathologists’ diagnostic impressions, including recognition of invasion, these subjective assessments have not been formally classified. Given that the tumor microenvironment is important in tumor growth, angiogenesis, and metastasis [2,3,4], and that stromal-epithelial interactions [5,6,7,8] contribute to progression of ductal carcinoma in situ to invasive breast cancer, we hypothesize that morphologic analysis of stroma could have importance in understanding breast carcinogenesis and diagnosis. This view is supported by evidence that the transition from ductal carcinoma in situ to invasion is characterized by greater changes in gene expression of stromal cells than in epithelial tumor cells [9, 10].
Apart from evaluation of lymphoid infiltrates, which are a diagnostic feature of medullary carcinoma and can be graded [11], stromal alterations are often subtle and difficult to characterize and quantify by light microscopy alone. Emerging data suggest that automated pattern recognition systems could be used to characterize stromal changes. For example, in a computer-generated automated analysis of routinely prepared hematoxylin and eosin (H&E) stained breast cancer tissue sections, Beck et al. [12] reported that stromal features were associated with breast cancer survival, and were more predictive of prognosis than epithelial features. Development of an automated computerized tool to identify and characterize tumor-associated stroma could have utility in pathologic diagnosis with respect to evaluating tumor margins and cancer field effects or in predicting the potential of ductal carcinoma in situ to progress to invasion, if occult neoplastic cells persist after treatment.
Development of robust computerized algorithms for discriminating patterns of normal stroma and tumor-associated stroma in histopathology images is a complex task, partly because validated morphologic criteria for distinguishing tumor-associated stroma are undefined. Machine learning approaches, and more specifically deep learning algorithms, could prove very suitable for accomplishing this objective as they are capable of learning the most discriminative features directly from a large set of classified diagnostic images, and therefore, do not require pre-defined morphologic criteria [13,14,15]. Thus, in a study of women who underwent image-guided breast biopsy to investigate a radiologic abnormality [16], we aimed to develop a clinically applicable algorithm reflecting the nature of breast biopsy specimens that pathologists receive for diagnosis. The objectives of the present study were: (1) to generate a deep learning algorithm that can identify and distinguish tumor-associated stromal alterations from stroma associated with benign breast disease in H&E stained sections of breast biopsies; and (2) to apply the deep learning algorithm to assess stromal characteristics in varying grades of ductal carcinoma in situ, which may represent a proxy for risk of invasion.
Materials and methods
Case selection
This analysis included 882 women, ages 40–65 years, referred for diagnostic image-guided breast biopsies (including ultrasound-guided needle core biopsy and stereotactic vacuum-assisted biopsy), and who participated in the Breast Radiology Evaluation and Study of Tissues (BREAST) Stamp Project [16] undertaken between 2007 and 2010 at the University of Vermont Larner College of Medicine and the University of Vermont Medical Center. Women provided informed consent, which included access to medical records, self-reported breast cancer risk information, blood and saliva donations, access to radiological images and pathological tissues for research and follow-up. The study was approved by appropriate ethics review boards at the University of Vermont and at the National Cancer Institute (National Institutes of Health).
Breast biopsies were performed as ultrasound-guided core needle biopsies (14-gauge) or as stereotactically-guided vacuum-assisted biopsies (9-gauge) that were routinely fixed in formalin, prepared as paraffin-embedded tissue sections, and stained with H&E for diagnosis. For study purposes, biopsies were classified as non-proliferative benign breast disease, proliferative benign breast disease without atypia, atypical hyperplasia, ductal or lobular carcinoma in situ or invasive carcinoma [17]. When biopsies included multiple tissue blocks, reflecting target and surrounding non-target tissues, we attempted to collect sections from both types of blocks, yielding a total of 2387 total H&E stained sections that were scanned at 20× (Aperio, ScanScope CS or Hamamatsu) as digital images (resulting specimen level pixel size 0.455 µm × 0.455 µm).
Reference standard
We manually annotated tissue structures to train our deep learning algorithms. We annotated breast tissue components such as stroma, epithelium, and fat, and also stromal regions in benign biopsies and adjacent to invasive cancer in the whole-slide images. Our analysis focused on the extracellular matrix stroma rather than focusing on specific features of stromal composition; thus, we analyzed all stromal areas in specific diagnostic contexts (benign, ductal carcinoma in situ and cancer) and in topographical proximity to diagnostic lesions. To analyze the pattern of stroma surrounding ductal carcinoma in situ lesions, the whole-slide images containing only ductal carcinoma in situ, and whole-slide images containing ductal carcinoma in situ with concurrent invasive cancer were annotated by a pathologist (MES). For each case, a subset of ducts containing ductal carcinoma in situ lesions was annotated on whole-slide images with point annotations in the center of the lesion and graded using standard criteria based on nuclear size and appearance, mitoses and detection of necrosis [18]. Ductal carcinoma in situ lesions in slides with concurrent invasive cancer were annotated if they were peripheral to the invasive component and its associated stroma.
Deep learning algorithms
Deep learning is a subfield of machine learning, where very general algorithms learn features directly from data for prediction and classification. Our whole-slide image classification system is based on multiple deep convolutional neural networks [13]. To enable assessment of unbiased performance of our algorithm, the dataset was randomly split into a training set containing 62% of the whole-slide images (1459 whole-slide images from 552 patients) and a testing set with the remaining slides (928 whole-slide images from 330 patients; Table 1).
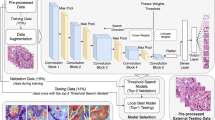
Briefly, using representative input patches from the annotated areas, a convolutional neural network model denoted ‘Network I’ was trained using the approach we previously described [19], to classify fat, stroma, and epithelium. Next, a second model “Network II” was trained operating on stromal regions recognized by Network I. Network II generated a probability that an image represented cancer-associated stroma. This model was trained using manually identified regions of stroma adjacent to invasive cancer and stroma in whole-slide images not containing tumor. Examples of ductal carcinoma in situ-associated stroma were not used in the training phase. To classify whole-slide images into normal/benign vs invasive cancer, a third model ‘Network III’ was constructed and composed of a small convolutional neural network stacked on top of Network II. Network III was trained to generate a score for the entire whole-slide image indicating the probability that the slide contained invasive cancer. More details about this network are described below. Figure 1 shows an overview of the entire classification system.
Overview of the system used for tumor-associated stroma identification and the system used for classification of the whole-slide image (WSI) into normal/benign or invasive cancer. a The top row shows the application of two convolutional neural networks (CNNs) for identifying regions of tumor-associated stroma in the WSI. CNN I classifies the tissue into epithelium, stroma, and fat. CNN II operates on stromal regions detected by CNN I and is trained to identify regions of tumor-associated stroma. b To classify the WSI into invasive cancer vs benign/normal, CNN III (stacked CNN) takes as input eight patches with high probabilities for tumor-associated stroma identified by CNN II and classifies the WSI by stacking a set of convolutional and fully connected layers on top of CNN II (see Table 1 in the supplement for details)
Network I and Network II in this study possess a VGG-Net-like architecture [20]. VGG-Net is a neural network architecture developed by Oxford’s Visual Geometry Group (VGG), which won the 2014 ImageNet Large Scale Visual Recognition Challenge 2014 task for object localization [21]. Details of our network configuration and training procedures are presented in Supplementary methods sections “Convolutional neural network architecture”, “Preprocessing of whole-slide images and ground truth ROIs” and “Training procedure”.
Description of convolutional neural network III for the classification of whole-slide images into normal/benign vs invasive cancer
To illustrate the potential of stroma characterization, Network III was constructed to identify cancerous biopsies based on the output of Network II only. The output feature map of the penultimate layer of Network II (the hidden layer whose output is fed to the final classification layer) is a compact representation of the input stromal image. Network III takes as input the feature maps from Network II for eight non-overlapping stromal tissue regions (size 152.9 µm × 152.9 µm), which were identified by Network II as harboring the strongest tumor-associated alterations and predicted the whole-slide image diagnosis (normal/benign vs invasive breast cancer; Fig. 1b). Details of the procedure for selecting those regions can be found in Supplementary methods sections “Selection of candidate stromal regions as input to convolutional neural network III”.
To generate the final score for each slide representing the probability of being a cancerous biopsy, we used an ensemble of two networks comprising Network III and a modified version of Network III without the last two fully connected layers. The average probability of the two networks was taken as the final score.
We additionally compared the performance of Network III, developed in the present study, with our recently published approach for whole-slide image classification [19]. Our previous system derived a total of 71 features from the outputs of both Network I and Network II and used these as input for a random forests classifier [22]. These features include the global tissue amount for epithelium, stroma, and fat as well as morphological features of epithelial areas and the spatial distribution of epithelial areas in the whole-slide image derived from two region adjacency graphs: Delaunay triangulation and area-Voronoi diagram [23].
Experiments
Classification of breast tissue whole-slide images as invasive carcinoma versus benign breast disease
The training data (1459 whole-slide images from 552 patients) used for this classification task was further divided into two sets, a preliminary set to define parameters, and a second (validation) set (comprising 10% of slides) that was used to perform final model selection and hyper-parameter optimization. The performance of our model was evaluated on the independent test set (928 whole-slide images from 330 patients) described previously above.
Analysis of ductal carcinoma in situ-associated stroma
In this experiment, we analyzed the stromal patterns surrounding ductal carcinoma in situ lesions on breast cancer slides. The ductal carcinoma in situ-associated stroma was analyzed using Network II which was trained to discriminate between normal and tumor-associated stroma. We first classified all the stromal pixels adjacent to annotated ductal carcinoma in situ lesions using Network II. Subsequently, we extracted two measures to quantify ductal carcinoma in situ-associated stroma. These measures are the mean and standard deviation of all tumor-associated stroma probabilities for the pixels surrounding ductal carcinoma in situ lesions. They were computed for a range of distances from the lesion’s margin. This analysis was performed independently on test slides with ductal carcinoma in situ lesions only and with test slides containing ductal carcinoma in situ accompanied by invasive cancer.
Statistical analysis
The area under the receiver operator characteristic curve was used to evaluate the performance of the system in discriminating between invasive carcinoma and benign breast disease biopsies. The receiver operator characteristics curve plots the sensitivity versus the false positive fraction (1-specificity) [24]. The area under the receiver operator characteristics curve ranges from 0-1 (with 1.0 representing the perfect classifier, 0.5 = to chance). 95% confidence intervals for the receiver operator characteristics curves were obtained using the percentile bootstrap method [25].
The significance test for comparing two correlated receiver operator characteristics curves, when comparing the performance of the proposed system for classification of whole-slide images with our previously described system, was done using the bootstrap method in R package “pROC” [26]. This method is based on the approach described by Hanley and McNeil [27] that takes into account the correlation that is induced by the paired nature of the data.
The one-way analysis of variance (ANOVA) and the Tukey post hoc test were used to compare the computed stromal measures described in results section “Analysis of ductal carcinoma in situ-associated stroma” for the patients with different ductal carcinoma in situ grades. A p-value < 0.05 was considered significant. All analyses were two-tailed.
Results
Classification of breast tissue whole-slide images as invasive carcinoma versus benign breast disease
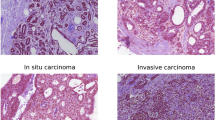
Network I, subdividing whole-slide images into regions consisting of epithelium, stroma, and fat achieved a pixel-level 3-class classification accuracy of 95.5% compared to reference standard, computed on a balanced subset of annotated pixels in the independent test set. Representative examples of tissue classification results are shown in Figure 2. Network II, used for classifying stroma into normal stroma and tumor-associated stroma, achieved a binary classification accuracy of 92.0% compared to reference standard, computed on a balanced subset of annotated pixels in the independent test set. Figure 2 shows the representative output probability map for a slide containing invasive cancer (Figs. 2a–c) and a normal slide (Figs. 2d–f).
Example results for the epithelium, stroma, and fat classification and tumor-stroma identification. a Shows a representative whole-slide image containing invasive cancer. b The classification map of epithelium (red), stroma (orange), and fat (green) for the image (a), produced by the first convolutional neural network model (CNN I). c The map produced by CNN II showing the tumor-stroma probability for the image (a), where green, yellow, and red represent low, medium and high probabilities, respectively. d A whole-slide image with benign epithelium, mostly terminal duct lobular units. e and f show the results for the epithelium, stroma, and fat classification and tumor-stroma identification for the image (d), respectively.
Figure 3 shows the receiver operator characteristics curves for the whole-slide image classification of invasive cancer vs. non-cancer using our proposed system and our previously published method [19]. Our newly developed convolutional neural network model achieved an area under the receiver operator characteristics curve of 0.962 (95% CI, 0.936–0.983), which was slightly higher (but not statistically significantly, p = 0.48) than our previously described approach [19], which achieved an area under the receiver operator characteristics curve of 0.948 (95% CI, 0.915–0.977).
Receiver operator characteristics curves with 95% confidence interval of the proposed system and the previously described approach [19] for the whole-slide image classification task of identifying invasive cancer vs. benign breast biopsies. 95% confidence intervals (CI) for the receiver operator characteristics curves were obtained using the percentile bootstrap method [25]. AUC denotes the area under the receiver operator characteristics curve.
In a subjective post hoc review, our study pathologist (MES) reviewed all misclassified cases: benign biopsies for which the algorithm score indicated a high cancer probability (“false positives”: probability > 0.70, n = 23), and invasive carcinomas for which the algorithm yielded a low probability of cancer (“false negatives”: probability < 0.27, n = 6). In addition, our pathologist reviewed a randomly selected group of correctly classified (true negative and true positive) cases within each diagnostic category. Given that we do not know what specific stromal features may be driving the machine classifier, all cases were reviewed in an un-masked fashion. The 23 benign biopsies that were misclassified as probably cancer, included 10 (43%) diagnoses of sclerosing adenosis; two (9%) of atypical hyperplasia and 11 (48%) of non-proliferative benign breast disease. Many of these benign specimens showed reactive stroma; these were evident in sclerosing adenosis as part of diagnostic criteria for this entity, but other benign samples showed reactions in relation to fat necrosis, suggestive of prior biopsy or ruptured ducts. The six biopsies of invasive carcinoma misclassified by the algorithm as probably benign contained minimal stroma, which likely reflects the combined effects of targeting the hypercellular center of the lesion and the small size of the biopsies.
Analysis of ductal carcinoma in situ-associated stroma
Figure 4 shows representative ductal carcinoma in situ lesions with different histological grades and their corresponding probability maps for tumor-associated stroma. Figure 5 shows examples of stroma patches for different grades of ductal carcinoma in situ harboring tumor-associated alterations. Boxplots for the mean and standard deviations of ductal carcinoma in situ-associated stroma probabilities for the pixels ≤ 175 µm from the ductal carcinoma in situ margin are shown in Figure 6a, b. In Figure 6a, b, each point is a single ductal carcinoma in situ lesion. Overall, the amount of tumor-associated stroma increased with increasing lesion grade. In addition, ductal carcinoma in situ lesions in slides without an invasive component presented less tumor-associated stroma compared to ductal carcinoma in situ lesions in slides demonstrating ductal carcinoma in situ with invasive cancer. Figures 6c, d show similar boxplots at the patient level where the average tumor stroma probability is shown as the mean of the scores of the highest grade foci per patient. A statistically significant difference was observed for the patient level means and standard deviations among the patients with different ductal carcinoma in situ grades (p = 0.023 and 0.005, respectively; one-way ANOVA). For slides containing ductal carcinoma in situ only, average tumor stromal probabilities were higher for higher grade ductal carcinoma in situ lesions, but this relationship was not evident for ductal carcinoma in situ-associated with invasive cancer. Values for tumor stromal probabilities showed greater variability for higher grade lesions. After multiple comparisons adjustment, the mean ductal carcinoma in situ-associated stroma probabilities (Fig. 6c) were significantly different between ductal carcinoma in situ grades 1 and 3 (p = 0.028). Mean differences between grades 2 and 3 and grades 1 and 2 were not statistically significant (p = 0.217 and p = 0.329). For the standard deviation of ductal carcinoma in situ-associated stroma probabilities (Fig. 6d), we observed statistically significant differences between grades 1 and 3 as well as grades 2 and 3 (p = 0.021 and p = 0.028). No statistical significance in standard deviation was observed between grades 1 and 2 (p = 0.619).
Example ductal carcinoma in situ lesions with different histological grades and their corresponding probability maps generated by convolutional neural network model II for identification of regions of tumor-associated stroma. The ductal carcinoma in situ lesions shown in a, b, and c have histological grades of 1, 2 and 3 (with necrosis), respectively. The tumor-stroma probability maps (green, yellow, and red represent low, medium and high probabilities) overlaid on the original images are shown for the pixels with distances below 175 µm from the ductal carcinoma in situ margin.
Examples of stroma patches for different grades of ductal carcinoma in situ with low and high probabilities for tumor-associated stroma. The first three rows show image patches (shown in green bounding boxes) for different grades of ductal carcinoma in situ that have low probabilities for tumor-associated stroma. The image patches in rows four to six (shown in red bounding boxes) show example patches for different grades of ductal carcinoma in situ with high probabilities for tumor-associated stroma. Note that each patch is taken from an independent ductal carcinoma in situ lesion; however, due to the lack of ductal carcinoma in situ grade 1 lesions with high tumor-stroma probabilities, the examples in the fourth row are taken from two lesions only. Subjectively, cellularity generally appears higher in grade 2 and 3 lesions.
Boxplots for the mean and standard deviation of tumor-associated stroma probabilities for the pixels at a distance of ≤175 µm from the ductal carcinoma in situ margin for different histological grades of ductal carcinoma in situ. a and b show the mean and standard deviation of tumor-associated stroma probabilities at the lesion level. The orange boxplots show the results for all ductal carcinoma in situ foci in slides diagnosed as ductal carcinoma in situ (n = 375). The green boxplots show the results for slides containing ductal carcinoma in situ accompanied by invasive cancer (n = 37). c and d show the mean and standard deviation of tumor-associated stroma probabilities at the patient level (n = 58) for slides with ductal carcinoma in situ diagnosis.
Figures 7a, b show the mean and standard deviation of ductal carcinoma in situ-associated stroma for rims of stroma at different distances from the ductal carcinoma in situ perimeter. As the distance from the ductal carcinoma in situ margin increases, the mean of the ductal carcinoma in situ-associated stroma probabilities decreases, but only slightly, and curves for different grades of ductal carcinoma in situ remain parallel up to 500 um from lesion periphery.
Plots of the mean (a) and standard deviation (b) of tumor-associated stroma probabilities for different ranges of distances from the ductal carcinoma in situ margin for different histological grades. For each measure (mean and standard deviation of probabilities), the curve for each grade is constructed using the mean along with the 95% confidence intervals around the mean of the population.
Discussion
In this study, we developed a state-of-the-art deep convolutional neural network for distinguishing benign breast disease from invasive breast cancer based on the identification and characterization of tumor-associated stromal alterations. In an independent test set, classification of breast biopsies as benign or malignant based solely on convolutional neural network analysis of stroma achieved an impressive area under the receiver operator characteristics curve of 0.962, consistent with highly accurate discrimination. A subjective post hoc review suggested that false positive results were associated with sclerosing adenosis, a benign lesion often associated with stromal changes, and with fat necrosis, whereas false negative specimens often demonstrated tightly packed cancer cells with minimal intervening stroma for evaluation. Without training the convolutional neural network on ductal carcinoma in situ lesions, we subsequently assessed whether tumor-associated stroma could be identified in tissues surrounding ductal carcinoma in situ and whether its extent varied with clinically important pathologic features. We detected greater amounts of tumor-associated stroma in grade 3 versus grade 1 ductal carcinoma in situ and also found that ductal carcinoma in situ-associated with an invasive component generally possessed higher amounts of tumor-associated stroma compared to slides containing ductal carcinoma in situ only. Thus, our work provides support for including morphological analysis of breast stroma in studies aiming to understand risk of ductal carcinoma in situ progressing to invasion and in defining the biology of invasive breast cancer.
To date, most previous work [28,29,30,31,32,33] using automated image analysis approaches to detect and classify breast cancer in histological images involved assessment of the morphology and arrangement of epithelial structures (e.g. nuclei, ducts). Generally, the aim of this research was to objectify, standardize and quantify features that are already appreciated as important by pathologists. Although subjective evaluation of stroma may provide cues that pathologists use in the histopathologic diagnosis of breast lesions, stroma is difficult to assess microscopically, and formal criteria for classifying stromal changes have not been developed and used clinically. Accordingly, agnostic approaches, such as using deep learning techniques, are well-suited to investigating the morphology of breast stroma because visual characterization or feature selection is not required.
In the surgical management of breast cancer, it may be important to excise malignant epithelium and tumor-associated stroma. The ability of the system to objectively identify regions of altered stroma associated with tumor may additionally complement the pathologist’s diagnosis and may assist in identifying stromal tissue that should be included in tumor margins.
A key goal of our project was to use an unbiased data-driven approach to examine potential relationships between the patterns of stroma surrounding ductal carcinoma in situ lesions and ductal carcinoma in situ grade. It is hypothesized that transformation of the stroma starts in an early phase of ductal carcinoma in situ development [5,6,7], and there is growing evidence that stroma contributes importantly to the transformation of ductal carcinoma in situ to invasion [5, 6, 9]. Thus, we tested the hypothesis that stromal alterations may serve as a proxy for the potential for ductal carcinoma in situ to undergo an invasive transformation.
Although we did not train our model on ductal carcinoma in situ, we found that tumor-associated stroma probabilities were significantly higher in grade 3 ductal carcinoma in situ, with the amount of tumor-associated stroma generally increasing with increasing lesion grade. Although we were unable to distinguish pathologically defined ductal carcinoma in situ grade 2 from ductal carcinoma in situ grade 1 or grade 3, data show that reproducibility of ductal carcinoma in situ grade 2 is poor [18], suggesting that this comparison may have limited value. Despite this limitation, data show that high-grade ductal carcinoma in situ may have a higher risk of recurrence after surgical excision than low-grade ductal carcinoma in situ, and when recurrences occur after ductal carcinoma in situ treatment [34,35,36], they occur earlier for higher grade lesions [36,37,38]. Studies also suggest that occult invasion is more common among women with image-guided biopsies diagnosed with higher grades of ductal carcinoma in situ [39, 40] and that this may be important because grade of invasive cancer is generally matched with grade of accompanying ductal carcinoma in situ [37, 41]. Further, a low percentage of high-grade ductal carcinoma in situ has been associated with positive axillary nodes or later metastases, suggesting that at least a subset of such lesions are associated with occult invasion or disseminate through an undefined mechanism. Finally, ongoing prospective trials (LORIS [42], LORD [43], and COMET [44]) are assessing conservative management of low-risk ductal carcinoma in situ, given indirect evidence that many such lesions will never cause harm during a woman’s lifetime. Our data suggest that consideration of evaluating stromal changes to assess its role as a potential biomarker of risk for recurrence may have value in such trials.
There are several limitations to our study. Our dataset was limited to one study population, thus repeating this analysis in other populations is important. Additionally, this study was limited to breast tissue sections obtained at time of biopsy and further insights might be obtained by assessing the stromal patterns on whole-slide images from subsequent matched breast tissue surgical resections. Although our comparison of tumor-associated stroma in pure ductal carcinoma in situ versus ductal carcinoma in situ with invasive cancer attempted to focus on areas of slides that were further away from invasive cancer, because of limited amount of tissue in some biopsies, there was a risk that stromal changes associated with some ductal carcinoma in situ areas reflected nearby invasion. Analyzing whole-slide images of resected specimens would help alleviate this risk, provided that avoiding changes associated with the prior biopsy site does not pose insurmountable challenges. Additionally, by virtue of the deep learning process, it is unclear what components of the stroma may be driving the machine classifier. Additional studies are needed to understand the biology of the stroma surrounding ductal carcinoma in situ, including the role of the vasculature, which we previously showed was increased in ductal carcinoma in situ versus benign biopsies from this study, with the highest microvessel density in invasive carcinoma [45]. Further experiments on larger cohorts of ductal carcinoma in situ with long-term clinical follow-up are needed, to study the potential that stromal features may have prognostic value. For example, stromal analysis may help define which ductal carcinoma in situ, grade 2, will behave indolently like grade 1 versus more aggressively like grade 3.
In conclusion, we have developed a deep learning approach utilizing convolutional neural network to identify the presence of cancer in whole-slide images based on tumor-associated stromal alterations in diagnostic image-guided breast biopsies. Further, we demonstrated that deep learning techniques can define stromal features that are related to ductal carcinoma in situ grade. Additional studies using these approaches with follow-up of ductal carcinoma in situ cases may be useful.
References
Dupont WD, Parl FF, Hartmann WH, et al. Breast cancer risk associated with proliferative breast disease and atypical hyperplasia. Cancer. 1993;71:1258–65.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37.
Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901.
Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52.
Provenzano PP, Eliceiri KW, Campbell JM, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38.
Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–48.
Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. The origin of the myofibroblasts in breast cancer. Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J Clin Invest. 1995;95:859–73.
Tlsty TD, Hein PW. Know thy neighbor: Stromal cells can contribute oncogenic signals. Curr Opin Genet Dev. 2001;11:54–9.
Ma X-J, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7.
Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchió C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–92.
Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–71.
Beck AH, Sangoi AR, Leung S, et al. Systematic Analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra13.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.
Litjens G, Kooi T, Ehteshami Bejnordi B, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210.
Gierach GL, Geller BM, Shepherd JA, et al. Comparison of mammographic density assessed as volumes and areas among women undergoing diagnostic image-guided breast biopsy. Cancer Epidemiol Biomark Prev. 2014;23:2338–48.
Gierach GL, Patel DA, Pfeiffer RM, et al. Relationship of terminal duct lobular unit involution of the breast with area and volume mammographic densities. Cancer Prev Res. 2016;9:149–58.
Pinder SE. Ductal carcinoma in situ (DCIS): Pathological features, differential diagnosis, prognostic factors and specimen evaluation. Mod Pathol. 2010;23:S8–S13.
Ehteshami Bejnordi B, Linz J, Glass B, et al. Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images. Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI), IEEE: Melbourne, VIC; 2017;929–32.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014:14091556.
Russakovsky O, Deng J, Su H, et al. ImageNet Large scale visual recognition challenge. Int J Comput Vision. 2015;115:211–52.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Okabe A, Boots B, Sugihara K. Spatial tessellations: concepts and applications of Voronoi diagrams. John Wiley & Sons, Inc., Chichester, New York, Brisbane, Toronto and Singapore; 1992. p. 532.
Glasziou P. The statistical evaluation of medical tests for classification and prediction. Stat Med. 2005;24:2582.
Efron B. Bootstrap methods: another look at the Jackknife. Ann Stat. 1979;7:1–26.
Robin X, Turck N, Hainard A, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:77.
Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–43.
Naik S, Doyle S, Agner S, et al. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. Proceedings of the 2008 IEEE 5th International Symposium on Biomedical Imaging (ISBI), IEEE: Paris, 2008;284–7.
Ehteshami Bejnordi B, Litjens G, Hermsen M, Karssemeijer N, A. W. M. van der Laak J. A multi-scale superpixel classification approach to the detection of regions of interest in whole slide histopathology images. Proceedings of the SPIE Medical Imaging 2015: Digital Pathology, SPIE Orlando, Florida; 2015;94200H.
Ehteshami Bejnordi B, Balkenhol M, Litjens G, et al. Automated detection of DCIS in whole-slide H&E stained breast histopathology images. IEEE Trans Med Imaging. 2016;35:2141–50.
Dundar MM, Badve S, Bilgin G, et al. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans Biomed Eng. 2011;58:1977–84.
Dong F, Irshad H, Oh E-Y, et al. Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLOS ONE. 2014;9:e114885.
Ehteshami Bejnordi B, Zuidhof G, Balkenhol M, et al. Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images. J Med Imaging. 2017;4:044504.
Solin LJ, Kurtz J, Fourquet A, et al. Fifteen-year results of breast-conserving surgery and definitive breast irradiation for the treatment of ductal carcinoma in situ of the breast. J Clin Oncol. 1996;14:754–63.
Silverstein MJ, Lagios MD, Groshen S, et al. The influence of margin width on local control of ductal carcinoma in situ of the breast. N Engl J Med. 1999;340:1455–61.
Benson JR, Jatoi I, Toi M. Treatment of low-risk ductal carcinoma in situ: is nothing better than something? Lancet Oncol. 2016;17:e442–e51.
Wallis MG, Clements K, Kearins O, et al. The effect of DCIS grade on rate, type and time to recurrence after 15 years of follow-up of screen-detected DCIS. Br J Cancer. 2012;106:1611–7.
Lagios MD, Margolin FR, Westdahl PR, Rose MR. Mammographically detected duct carcinoma in situ. Frequency of local recurrence following tylectomy and prognostic effect of nuclear grade on local recurrence. Cancer. 1989;63:618–24.
Bagnall MJC, Evans AJ, Wilson ARM, et al. Predicting invasion in mammographically detected microcalcification. Clin Radiol. 2001;56:828–32.
Silver SA, Tavassoli FA. Mammary ductal carcinoma in situ with microinvasion. Cancer. 1998;82:2382–90.
Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223:307–17.
Francis A, Bartlett J, Billingham L, et al. The LORIS trial: a multicentre, randomized phase III trial of standard surgery versus active monitoring in women with newly diagnosed low risk ductal carcinoma in situ. Cancer Res. 2013;73:OT2-3-01.
Elshof LE, Tryfonidis K, Slaets L, et al. Feasibility of a prospective, randomised, open-label, international multicentre, phase III, non-inferiority trial to assess the safety of active surveillance for low risk ductal carcinoma in situ—The LORD study. Eur J Cancer. 2015;51:1497–510.
Youngwirth L, Boughey J, Hwang E. Surgery versus monitoring and endocrine therapy for low-risk DCIS: The COMET Trial. Bull Am Coll Surg. 2017;102:62–3.
Felix AS, Lenz P, Pfeiffer RM, et al. Relationships between mammographic density, tissue microvessel density, and breast biopsy diagnosis. Breast Cancer Res. 2016;18:88.
Acknowledgements
This project was funded in part by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Bethesda, Maryland and a competitive award to MES and LAB funded through the sale of breast cancer awareness postage stamps. The authors wish to acknowledge the financial support by the European Union FP7 funded VPHPRISM project under the grant agreement n601040. Pamela Vacek and Donald Weaver are currently funded under a U01 exploring stromal contributions to tumor progression (U01 CA196383).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ehteshami Bejnordi, B., Mullooly, M., Pfeiffer, R.M. et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol 31, 1502–1512 (2018). https://doi.org/10.1038/s41379-018-0073-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-018-0073-z
This article is cited by
-
Progression from ductal carcinoma in situ to invasive breast cancer: molecular features and clinical significance
Signal Transduction and Targeted Therapy (2024)
-
Prognostic significance of collagen signatures at breast tumor boundary obtained by combining multiphoton imaging and imaging analysis
Cellular Oncology (2024)
-
Artificial intelligence in diagnostic pathology
Diagnostic Pathology (2023)
-
Biological insights and novel biomarker discovery through deep learning approaches in breast cancer histopathology
npj Breast Cancer (2023)
-
Heterogeneity of the tumor immune microenvironment and clinical interventions
Frontiers of Medicine (2023)