Abstract
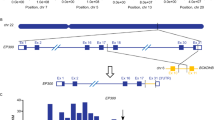
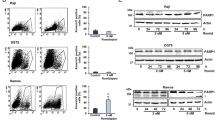
Somatic mutations affecting CREBBP and EP300 are a hallmark of diffuse large B-cell lymphoma (DLBCL). These mutations are frequently monoallelic, within the histone acetyltransferase (HAT) domain and usually mutually exclusive, suggesting that they might affect a common pathway, and their residual WT expression is required for cell survival. Using in vitro and in vivo models, we found that inhibition of CARM1 activity (CARM1i) slows DLBCL growth, and that the levels of sensitivity are positively correlated with the CREBBP/EP300 mutation load. Conversely, treatment of DLBCLs that do not have CREBBP/EP300 mutations with CARM1i and a CBP/p300 inhibitor revealed a strong synergistic effect. Our mechanistic data show that CARM1i further reduces the HAT activity of CBP genome wide and downregulates CBP-target genes in DLBCL cells, resulting in a synthetic lethality that leverages the mutational status of CREBBP/EP300 as a biomarker for the use of small-molecule inhibitors of CARM1 in DLBCL and other cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All raw and normalized RNA-Seq and ChIP-seq data that support the findings of this study have been deposited in the GEO SuperSeries under accession number GSE152668.
References
Al-Tourah AJ, Gill KK, Chhanabhai M, Hoskins PJ, Klasa RJ, Savage KJ, et al. Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. J Clin Oncol. 2008;26:5165–9.
Guo L, Lin P, Xiong H, Tu S, Chen G. Molecular heterogeneity in diffuse large B-cell lymphoma and its implications in clinical diagnosis and treatment. Biochim Biophys Acta. 2018;1869:85–96.
Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33.
MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–39.
Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57.
Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci USA. 2015;112:E1116–25.
Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109:3879–84.
Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95.
Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–7.
Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1398–403.
Ceschin DG, Walia M, Wenk SS, Duboe C, Gaudon C, Xiao Y, et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25:1132–46.
Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–66.
Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA. 2005;102:3611–6.
Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–11.
Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–6.
Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–23.
Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7.
Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72.
Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, et al. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29:3413–23.
Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA. 2003;100:6464–8.
Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, et al. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279:25339–44.
Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, et al. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–8.
Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9:47.
O’Brien KB, Alberich-Jorda M, Yadav N, Kocher O, Diruscio A, Ebralidze A, et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137:2147–56.
Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, et al. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285:1147–52.
Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50.
El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci USA. 2006;103:13351–6.
Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–9.
Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD, et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer 2010;10:197.
Osada S, Suzuki S, Yoshimi C, Matsumoto M, Shirai T, Takahashi S, et al. Elevated expression of coactivator-associated arginine methyltransferase 1 is associated with early hepatocarcinogenesis. Oncol Rep. 2013;30:1669–74.
Mann M, Cortez V, Vadlamudi R. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013;34:1468–75.
Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36.
Karakashev S, Zhu H, Wu S, Yokoyama Y, Bitler BG, Park PH, et al. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat Commun. 2018;9:631.
Bao J, Di Lorenzo A, Lin K, Lu Y, Zhong Y, Sebastian MM, et al. Mouse models of overexpression reveal distinct oncogenic roles for different type I protein arginine methyltransferases. Cancer Res. 2019;79:21–32.
Nakayama K, Szewczyk MM, Dela Sena C, Wu H, Dong A, Zeng H, et al. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9:18480–93.
Drew AE, Moradei O, Jacques SL, Rioux N, Boriack-Sjodin AP, Allain C, et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7:17993.
Greenblatt SM, Man N, Hamard PJ, Asai T, Karl D, Martinez C, et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell. 2018;33:1111–27.e5.
Cheng D, Vemulapalli V, Lu Y, Shen J, Aoyagi S, Fry CJ, et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance. 2018;1:e201800117.
Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–73.
Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7:322–37.
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–6.
Andersen CL, Asmar F, Klausen T, Hasselbalch H, Gronbaek K. Somatic mutations of the CREBBP and EP300 genes affect response to histone deacetylase inhibition in malignant DLBCL clones. Leuk Res Rep. 2012;2:1–3.
Hashwah H, Schmid CA, Kasser S, Bertram K, Stelling A, Manz MG, et al. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc Natl Acad Sci USA. 2017;114:9701–6.
Haery L, Lugo-Pico JG, Henry RA, Andrews AJ, Gilmore TD. Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth. Mol Cancer. 2014;13:29.
Hammitzsch A, Tallant C, Fedorov O, O’Mahony A, Brennan PE, Hay DA, et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci USA. 2015;112:10768–73.
Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–74.
Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7:38–53.
Meyer SN, Scuoppo C, Vlasevska S, Bal E, Holmes AB, Holloman M, et al. Unique and shared epigenetic programs of the CREBBP and EP300 acetyltransferases in germinal center B cells reveal targetable dependencies in lymphoma. Immunity. 2019;51:535–47. e9.
Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809.
Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006;107:4407–16.
Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–7.
Kaushik S, Liu F, Veazey KJ, Gao G, Das P, Neves LF, et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 2018;32:499–509.
Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30:789–99.
Lu X, Fernando TM, Lossos C, Yusufova N, Liu F, Fontan L, et al. PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood. 2018;132:2026–39.
George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53.
Kishimoto M, Kohno T, Okudela K, Otsuka A, Sasaki H, Tanabe C, et al. Mutations and deletions of the CBP gene in human lung cancer. Clin Cancer Res. 2005;11:512–9.
Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8.
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–9.
Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–81.
Wilson BG, Helming KC, Wang X, Kim Y, Vazquez F, Jagani Z, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34:1136–44.
Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10.
So CK, Nie Y, Song Y, Yang GY, Chen S, Wei C, et al. Loss of heterozygosity and internal tandem duplication mutations of the CBP gene are frequent events in human esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:19–27.
Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, et al. Targeting p300 addiction in CBP-Deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–45.
Gao G, Zhang L, Villarreal OD, He W, Su D, Bedford E, et al. PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res. 2019;47:5038–48.
Acknowledgements
We thank Manu Sebastian Ph.D., Abhinav Jain Ph.D., Michelle Barton Ph.D., Carlos Perez, and members of the MAS laboratory for technical support and helpful discussions. Core facilities were supported by the NIH Grant P30CA16672 and Cancer Prevention Research Institute of Texas (CPRIT) Grants RP120348, RP170002, and RP170628. This work was supported by CPRIT Recruitment of First-time Tenure-Track Faculty award RR160097 (to HX), NIH grant GM126421 (to MTB), Andrew Sabin Family Fellow Award, American Society of Hematology Junior Faculty Scholar Award, and CPRIT First-time Tenure-Track Faculty award RR150039 (to MAS). MAS and HX are CPRIT scholars in cancer research.
Author information
Authors and Affiliations
Contributions
KJV, MG, HX, YL, MTB, and MAS participated in the study design. KJV, DC, GG, MP-O, and HTV participated in cell culture experiments, in vitro drug treatments, protein, and mRNA quantifications, and analyzed data. KJV performed and analyzed all ChIP-qPCR experiments. KJV and SAS performed mouse breeding, xenografts, and in vivo drug treatments, and analyzed data. OV performed the synergistic drug computational analysis. KJV, KL and YL performed all the RNA-seq and ChIP-seq computational analysis. KJV and MAS wrote the paper and all authors reviewed it. MAS supervised the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Veazey, K.J., Cheng, D., Lin, K. et al. CARM1 inhibition reduces histone acetyltransferase activity causing synthetic lethality in CREBBP/EP300-mutated lymphomas. Leukemia 34, 3269–3285 (2020). https://doi.org/10.1038/s41375-020-0908-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-020-0908-8
This article is cited by
-
The emerging role of CARM1 in cancer
Cellular Oncology (2024)
-
Histone regulator KAT2A acts as a potential biomarker related to tumor microenvironment and prognosis of diffuse large B cell lymphoma
BMC Cancer (2023)
-
The NFIB/CARM1 partnership is a driver in preclinical models of small cell lung cancer
Nature Communications (2023)
-
CARM1 promotes gastric cancer progression by regulating TFE3 mediated autophagy enhancement through the cytoplasmic AMPK-mTOR and nuclear AMPK-CARM1-TFE3 signaling pathways
Cancer Cell International (2022)
-
Systematic pan-cancer landscape identifies CARM1 as a potential prognostic and immunological biomarker
BMC Genomic Data (2022)