Abstract
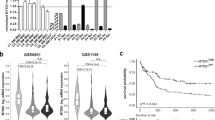
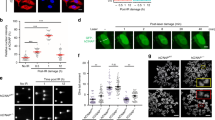
The transcriptional regulator far upstream element binding protein 1 (FUBP1) acts as an oncoprotein in solid tumor entities and plays a role in the maintenance of hematopoietic stem cells. However, its potential function in leukemia is unknown. In murine models of chronic (CML) and acute myeloid leukemia (AML) induced by BCR-ABL1 and MLL-AF9, respectively, knockdown of Fubp1 resulted in prolonged survival, decreased numbers of CML progenitor cells, decreased cell cycle activity and increased apoptosis. Knockdown of FUBP1 in CML and AML cell lines recapitulated these findings and revealed enhanced DNA damage compared to leukemia cells expressing wild type FUBP1 levels. FUBP1 was more highly expressed in human CML compared to normal bone marrow cells and its expression correlated with disease progression. In AML, higher FUBP1 expression in patient leukemia cells was observed with a trend toward correlation with shorter overall survival. Treatment of mice with AML with irinotecan, known to inhibit topoisomerase I and FUBP1, significantly prolonged survival alone or in combination with cytarabine. In summary, our data suggest that FUBP1 acts as cell cycle regulator and apoptosis inhibitor in leukemia. We demonstrated that FUBP1 might play a role in DNA repair, and its inhibition may improve outcome in leukemia patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538–45.
Zhang J, Chen QM. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32:2907–16.
Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–66.
Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–80.
Rabenhorst U, Beinoraviciute-Kellner R, Brezniceanu ML, Joos S, Devens F, Lichter P, et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology. 2009;50:1121–9.
Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30:846–58.
Malz M, Weber A, Singer S, Riehmer V, Bissinger M, Riener MO, et al. Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology. 2009;50:1130–9.
Jia MY, Wang YJ. Far upstream element-binding protein 1 (FUBP1) expression differs between human colorectal cancer and non-cancerous tissue. Neoplasma. 2014;61:533–40.
Zhang F, Tian Q, Wang Y. Far upstream element-binding protein 1 (FUBP1) is overexpressed in human gastric cancer tissue compared to non-cancerous tissue. Onkologie. 2013;36:650–5.
Liu ZH, Hu JL, Liang JZ, Zhou AJ, Li MZ, Yan SM, et al. Far upstream element-binding protein 1 is a prognostic biomarker and promotes nasopharyngeal carcinoma progression. Cell Death Dis. 2015;6:e1920.
Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008;8:369.
Duan J, Bao X, Ma X, Zhang Y, Ni D, Wang H, et al. Upregulation of far upstream element-binding protein 1 (FUBP1) promotes tumor proliferation and tumorigenesis of clear cell renal cell carcinoma. PLoS ONE 2017;12:e0169852.
Baumgarten P, Harter PN, Tonjes M, Capper D, Blank AE, Sahm F, et al. Loss of FUBP1 expression in gliomas predicts FUBP1 mutation and is associated with oligodendroglial differentiation, IDH1 mutation and 1p/19q loss of heterozygosity. Neuropathol Appl Neurobiol. 2014;40:205–16.
Singer S, Malz M, Herpel E, Warth A, Bissinger M, Keith M, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res. 2009;69:2234–43.
Rabenhorst U, Thalheimer FB, Gerlach K, Kijonka M, Bohm S, Krause DS, et al. Single-stranded DNA-binding transcriptional regulator FUBP1 is essential for fetal and adult hematopoietic stem cell self-renewal. Cell Rep. 2015;11:1847–55.
Khageh Hosseini S, Kolterer S, Steiner M, von Manstein V, Gerlach K, Trojan J, et al. Camptothecin and its analog SN-38, the active metabolite of irinotecan, inhibit binding of the transcriptional regulator and oncoprotein FUBP1 to its DNA target sequence FUSE. Biochem Pharmacol. 2017;146:53–62.
Dixit U, Liu Z, Pandey AK, Kothari R, Pandey VN. Fuse binding protein antagonizes the transcription activity of tumor suppressor protein p53. BMC Cancer. 2014;14:925.
Jacob AG, Singh RK, Mohammad F, Bebee TW, Chandler DS. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J Biol Chem. 2014;289:17350–64.
Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–22.
Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–7.
Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–412.
Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci USA. 2006;103:16870–5.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Johnson FM, Krug LM, Tran HT, Shoaf S, Prieto VG, Tamboli P, et al. Phase I studies of imatinib mesylate combined with cisplatin and irinotecan in patients with small cell lung carcinoma. Cancer. 2006;106:366–74.
He L, Liu J, Collins I, Sanford S, O’Connell B, Benham CJ, et al. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–44.
Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607.
Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D, et al. MYC deregulation in primary human cancers. Genes (Basel). 2017;8:pii: E151.
Abraham SA, Hopcroft LE, Carrick E, Drotar ME, Dunn K, Williamson AJ, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–6.
Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, NS W, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553:515–20.
Debaize L, Jakobczyk H, Avner S, Gaudichon J, Rio A-G, Sérandour AA, et al. Interplay between transcription regulators RUNX1 and FUBP1 activates an enhancer of the oncogene c-KIT and amplifies cell proliferation Nucleic acids research. 2018;46:11214–28.
Malz M, Bovet M, Samarin J, Rabenhorst U, Sticht C, Bissinger M, et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60:1241–50.
Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q, Chung MC. 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics. 2008;8:5086–96.
de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, et al. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J Proteom. 2014;99:26–39.
Humar B, Graziano F, Cascinu S, Catalano V, Ruzzo AM, Magnani M, et al. Association of CDH1 haplotypes with susceptibility to sporadic diffuse gastric cancer. Oncogene. 2002;21:8192–5.
Tang Q, Xia W, Ji Q, Ni R, Bai J, Li L, et al. Role of far upstream element binding protein 1 in colonic epithelial disruption during dextran sulphate sodium-induced murine colitis. Int J Clin Exp Pathol. 2014;7:2019–31.
Lasserre JP, Fack F, Revets D, Planchon S, Renaut J, Hoffmann L, et al. Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J Proteome Res. 2009;8:5485–96.
Xu SG, Yan PJ, Shao ZM. Differential proteomic analysis of a highly metastatic variant of human breast cancer cells using two-dimensional differential gel electrophoresis. J Cancer Res Clin Oncol. 2010;136:1545–56.
Ding Z, Liu X, Liu Y, Zhang J, Huang X, Yang X, et al. Expression of far upstream element (FUSE) binding protein 1 in human glioma is correlated with c-Myc and cell proliferation. Mol Carcinog. 2015;54:405–15.
Minderman H, O’Loughlin KL, Smith PF, Pendyala L, Greco WR, Sweeney KG, et al. Sequential administration of irinotecan and cytarabine in the treatment of relapsed and refractory acute myeloid leukemia. Cancer Chemother Pharmacol. 2006;57:73–83.
Acknowledgements
This work was supported by grant 2015.039.1 to MZ and DSK from the Wilhelm Sander-Stiftung.We thank the Serviceeinrichtung Transkriptomanalyselabor (TAL) in Göttingen, Germany, for performing the RNA sequencing work.
Author contributions
VTH designed and carried out experiments, analyzed the data and wrote a first draft of the manuscript. DV designed and carried out experiments and analyzed the data. SG, PL, MS, KG, EW and UM-K assisted with experiments. BM and HF assisted with the analysis of the RNA-sequencing data. HB, TO, and AW provided and analyzed the human immunohistochemical data. JMV and VGO provided and analyzed the human genomic datasets. MZ and DSK designed experiments, supervised the project and analyzed data. DSK wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hoang, V.T., Verma, D., Godavarthy, P.S. et al. The transcriptional regulator FUBP1 influences disease outcome in murine and human myeloid leukemia. Leukemia 33, 1700–1712 (2019). https://doi.org/10.1038/s41375-018-0358-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0358-8
This article is cited by
-
LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC
Journal of Hematology & Oncology (2021)
-
The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC
Molecular Cancer (2021)
-
Human Adipose-Derived Mesenchymal Stromal Cells Exhibit High HLA-DR Levels and Altered Cellular Characteristics under a Xeno-free and Serum-free Condition
Stem Cell Reviews and Reports (2021)