Abstract
Electron beam (EB) irradiation is useful to reduce the recurrence of keloids; however, the underlying mechanism remains unknown. MicroRNA-21 (miR-21), which regulates autophagy during cancer radiation therapy, was identified as a potential therapeutic target for keloids. Here, we investigate the regulatory mechanism(s) of miR-21-5p on keloid fibroblast autophagy and migration after EB irradiation. The microRNA expression profile of the keloid dermis was examined by performing a microRNA microarray. Levels of LC3B and Beclin-1 were detected by immunohistochemical and western blot analysis in the keloid dermis and fibroblasts. Autophagy and apoptosis were tested in keloid fibroblasts after EB irradiation or transfection with an miR-21-5p inhibitor using electron microscopy, a Cyto-ID Green Autophagy Detection Kit, and an Annexin V PE Apoptosis Detection Kit. Migration was analyzed by an in vitro scratch–wound healing assay. Mechanistic tests were performed using small interfering RNAs to phosphatase and tensin homolog (siPTEN). Levels of miR-21-5p, PTEN, programmed cell death 4 (PDCD4), p-AKT, and apoptosis- and autophagy-associated genes were examined by qRT-PCR and western blotting. LC3B expression and migration ability were enhanced in fibroblasts and the keloid margin dermis compared with those in the adjacent normal skin. Both EB irradiation and an miR-21-5p inhibitor reduced keloid fibroblast autophagy, which was accompanied by decreased expression of miR-21-5p, p-AKT, and LC3B-II and increased expression of PTEN, PDCD4, and apoptosis-related genes. MiR-21-5p downregulation inhibited migration and suppressed LC3B expression and this was reversed by PTEN reduction. In conclusion, with increasing apoptosis, EB irradiation inhibits autophagy in keloid fibroblasts by reducing miR-21-5p, which regulates migration and LC3B expression via PTEN/AKT signaling. These data suggest a potential mechanism wherein miR-21-5p inhibition regulates autophagy and migration in EB-irradiated keloid fibroblasts, effectively preventing local invasion and recurrence. Therefore, miR-21-5p could be a new therapeutic target, to replace EB irradiation, and control keloid relapse.
Similar content being viewed by others
Introduction
Keloids are a fibroproliferative disorder in which scar tissue continues with tumor-like growth and extends beyond the margins of the initial insult [1]. No satisfactory treatment has been developed due to their high rate of recurrence (up to 70–80% post excision). The simple surgical excision of a keloid is associated with a worse scar that is even larger than the initial lesion due to the stimulation of fibroblast proliferation and additional collagen synthesis, prompting quick keloid relapse. Thus, surgical therapy should be combined with adjuvant treatment such as pressure, corticosteroids, and radiotherapy [2]. Keloids mainly involve hyperproliferative fibroblasts in which apoptosis is suppressed [3, 4], and active fibroblast migration in the margin area is involved in local invasion [5]. Postoperative adjuvant electron beam (EB) irradiation is a useful method to reduce keloid recurrence, lowering the relapse rate to 2.2–9.59%, but radiotherapy-related adverse effects include skin ulcerations, unhealed wounds, skin hyperpigmentation and/or telangiectasia with depigmentation, and the risk of radiation-induced malignancy, among others. Further, radiation therapy is contraindicated in children, as well as in areas of high carcinogenic potential such as the breast and thyroid [6, 7]. Previously, cDNA microarrays showed that proapoptotic genes in keloid fibroblasts are upregulated following EB irradiation exposure [8]. However, little is known regarding the molecular mechanism(s) through which EB irradiation suppresses keloid growth.
Autophagy is an intracellular process in which an altered membrane structure results in the packaging and delivery of organelles and macromolecules to lysosomes for degradation, promoting cytoplasmic component renewal. The autophagic pathway includes the initiation, elongation, and formation of autophagosomes and autolysosomes. Microtubule-associated protein 1 light chain 3 (LC3) is localized to the membrane bilayer of autophagosomes, and its conversion from the cytosolic form LC3-I to the autophagosome membrane-associated form LC3-II is considered a marker of autophagy [9]. This process plays a key role in diseases including cancer and fibrosis [10]. The expression of LC3B, an isoform of the LC3 family, is a common feature in cancers associated with metastasis [11]. Recent studies have shown that inhibiting autophagy reduces tumor cell invasion and migration, which is critical during the early stages of metastasis, including local invasion and intravasation [12]. Meanwhile, the upregulation of LC3 in keloids suggests that the inhibition of autophagy might also have therapeutic implications [13]. However, it is unknown whether autophagy is involved in local invasion and the response to EB irradiation in keloid fibroblasts.
MicroRNA-21 (miR-21) controls cellular apoptosis, autophagy, migration, and invasion [14]. It is also overexpressed in most cancers and various fibrotic diseases and is associated with metastasis and recurrence [15, 16]. Recent findings have suggested that miR-21 is a novel target for cancer radiation therapy due to its effects on apoptosis and autophagy [17]. MiR-21 is overexpressed in keloids, affecting the proliferation and apoptosis of keloid fibroblasts [18]. Therefore, the regulatory function of miR-21 in the autophagy and migration of keloid fibroblasts and in EB-irradiated keloid fibroblasts needs to be investigated.
In the current study, we investigated the expression of autophagy-associated marker genes in different areas of keloid tissues, changes in autophagy in keloid fibroblasts after EB irradiation, and the expression of miR-21-5p and apoptosis- and autophagy-related genes. In addition, the effects of miR-21-5p on regulating autophagy, migration, and apoptosis, as well as its influence on target genes and related signaling pathways, were examined. Our results provide a possible mechanism in which miR-21-5p inhibition regulates autophagy and migration in keloid fibroblasts after EB irradiation, effectively preventing local invasion and recurrence.
Materials and methods
Skin tissue specimens for microRNA microarray analysis and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Keloid samples were obtained from eight Chinese patients including six female and two male patients (19–30 years, mean 23.4 ± 3.7) who underwent surgical excision; this included five keloids from the earlobes and three from the trunks. Normal skin samples were obtained from the trunks of eight healthy donors during plastic surgery procedures, including two female and six male patients (8–48 years of age, mean 24.1 ± 11.5). Among them, the dermis samples from three keloid patients and three normal skin donors were detected using the microRNA microarray. All dermis samples from eight keloid patients and eight normal skin donors were used to validate the results of the microRNA microarray by qRT-PCR analysis. All experiments in our study were conducted in accordance with the Declaration of Helsinki and were approved by the Ethics Committee of the Plastic Surgery Hospital (Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) and informed consent was provided by all patients and donors.
MicroRNA microarray analysis and qRT-PCR
The dermis was separated from the keloid and normal skin samples after being incubated in 2.5 mg/mL dispase solution (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C for 16 h. Total RNA was extracted using a mirVana™ miR Isolation Kit (Ambion, Austin, TX, USA), and RNA integration was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Dermis samples from keloids and normal skin (n = 3 each) with an RNA integrity number ≥6.0 and 28S/18S ratio ≥0.7 were assessed using Agilent’s Human MicroRNA Microarray Release 18.0 and scanned with an Agilent Microarray Scanner (Agilent Technologies) at Shanghai Biochip Co., Ltd. (Shanghai, China). Raw data were normalized by the Quantile algorithm using Gene Spring Software 11.0. MiRs with a fold-change in expression (keloid/normal skin) <0.5 or >2 and a false discovery rate corrected p-value < 0.01 were considered down- or upregulated, respectively.
To validate miR microarray data, six miRs were selected for detection by SYBR Green qRT-PCR in keloid and normal dermis samples (n = 8 each). qRT-PCR was performed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) and Power SYBR Green PCR Master Mix with a 7900 HT Sequence Detection System (ABI, Foster, CA, USA). The comparative threshold cycle method was used to measure PCR amplification fold-differences. The expression of miRs was normalized to that of U6. Each experiment was performed in triplicate. All primers were synthesized by Invitrogen (Beijing, China). Primer sequences (5′-3′) were as follows: hsa-miR-21-5p: TAGCTTATCAGACTGATGTTGA; hsa-miR-214-5p: GCCTGTCTACACTTGCTGTGA; hsa-miR-542-5p: GGATCATCATGTCACGAGAAAA; hsa-miR-150-3p: CTGGTACAGGCCTGGGAAA; hsa-miR-493-5p: GTTGTACATGGTAGGCTTTCATTAA; hsa-miR-409-3p: GAATGTTGCTCGGTGAACC; and U6: TTCGTGAAGCGTTCCATATTTT.
Immunohistochemistry
Keloid tissues were fixed in 4% paraformaldehyde for 24 h, embedded in paraffin, and sectioned at 6 μm. Immunohistochemical studies were performed using standard procedures. Autophagy-related markers were detected using specific primary antibodies including rabbit polyclonal anti-LC3B (microtubule-associated protein 1 LC3 beta; 1:500, GeneTex, Inc. Irvine, CA, USA) and mouse monoclonal anti-Beclin-1 (1:500, Abcam, Cambridge, MA, US). Secondary antibodies from the Two-Step Polymer Non-Biotin HRP Detection System for DAB (Golden Bridge International, Mukilteo, WA, USA) were used. Stained immunohistochemical sections were imaged with a Leica DM3000 microscope (Leica Microsystems Gmbh, Wetzlar, Germany). Keloid samples were obtained from six Chinese patients, and the patient data are presented in Supplemental Table 1.
In vitro scratch–wound healing migration assays
Keloid and adjacent normal skin fibroblasts were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen Gibco, Carlsbad, CA, USA) and passaged at confluence with trypsin/EDTA (Thermo HyClone, South Logan, UT, USA). Fibroblasts at passages 3–5 from three independent keloid samples were used in our following experiments. The patient data regarding the sources of fibroblasts are provided in Supplemental Table 1.
Fibroblasts were seeded in 6-well plates, grown to confluency, and then scratched across the center using a 200-μL pipette tip; the media were then replenished with FBS-free DMEM to inhibit cell proliferation. Photographs were taken at the time of wounding (0 h) and at 48 h. Migration was quantified using Image J software. The data were expressed as a percentage of the wound healed area of the scratch filled by fibroblasts. The mean value was determined from triplicate experiments.
EB irradiation of keloid fibroblasts
Fibroblasts (2 × 104) were seeded in 25-cm2 flasks and cultured for 24 h before being exposed to single doses of 0-, 5-, 10-, 15-, 20-, or 30-Gy EB irradiation using a linear accelerator (Varian 600 C/D, Palo Alto, CA, USA) and cultured for 3 days. Apoptotic cells were detected using an Annexin V PE Apoptosis Detection Kit (eBioscience, San Diego, CA, USA) and Muse Cell Analyzer (Millipore, Hayward, CA, USA) according to the manufacturers’ protocols.
Autophagy detection
Autophagy induction was evaluated 1 h after subjecting cells to a single dose of 20-Gy EB irradiation by trypsinizing cells, fixing them in 2.5% glutaraldehyde in 0.1 M phosphate buffer, and then postfixing them in 1% osmium tetroxide buffer. After dehydration in a graded series of ethanol concentrations, cells were embedded and sectioned. The sectioned grids were stained with saturated solutions of uranyl acetate and lead citrate before being examined with an H-7650 electron microscope (Hitachi, Tokyo, Japan).
Cyto-ID Green Autophagy Detection staining was performed in accordance with the manufacturer’s instructions (Enzo Life Sciences, Farmingdale, NY, USA). Briefly, cells were grown to 70% confluence and transfected with a hsa-miR-21-5p inhibitor or corresponding inhibitor negative control at a final concentration of 25 nM (Ambion, Austin, TX, USA). After 48 h, cells were stained with Cyto-ID Green Detection Reagent and Hoechst 33342 nuclear dye solution at 37 °C for 30 min in the dark before being imaged with a Nikon TE2000 Microscope (Nikon, Tokyo, Japan). For flow cytometry, cells were digested and stained with the Cyto-ID Green Detection Reagent at 37 °C for 30 min and then analyzed with a FACSAria II flow cytometer (BD Biosciences, Mountain View, CA, USA).
MiR-21-5p antagonism and PTEN RNA interference
When keloid fibroblasts reached 80% confluence in 6-well plates, an hsa-miR-21-5p inhibitor or corresponding negative control (25 nM; Ambion) was transfected into separate cell populations using Lipofectamine RNAiMAX in OptiMEM I (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions.
The selected RNA duplex specific for PTEN mRNA (siPTEN, 60 nM) was transfected into keloid fibroblasts with Transfection Reagent in Transfection Medium (Santa Cruz Biotechnology, Santa Cruz, CA).
After transfecting fibroblasts in 6-well plates with an miR-21-5p inhibitor or negative control (25 nM) for 48 h or with siPTEN (60 nM) for 24 h with subsequent miR-21-5p inhibitor or negative control transfection (25 nM) for 48 h, total RNA and protein were simultaneously isolated from fibroblasts using a mirVana™ PARIS™ Kit (Ambion). Cells in each well of 6-well plates were seeded in two wells of 24-well plates for in vitro scratch–wound healing migration assays as described.
Quantitative reverse-transcription polymerase chain reaction
The expression of miR-21-5p was detected using a TaqMan MicroRNA Assay System and TaqMan MicroRNA Reverse Transcription Kit (ABI) in a DNA Engine Peltier Thermal Cycler (Bio-Rad) following the manufacturer’s instructions. Quantitative PCR was performed using the TaqMan Universal Master Mix II (ABI) in a LightCycler 480 II Real-Time PCR System (Roche, Rotkreuz, Switzerland). U6 expression was used to normalize miR-21-5p expression. Each experiment was performed in triplicate.
qRT-PCR of mRNAs was performed using a FastStart Universal SYBR Green Master Kit (ROX) (Roche Applied Science, Mannheim, Germany) and LightCycler 480 II Real-Time PCR System (Roche) according to the manufacturer’s instructions. The comparative threshold cycle method was used to measure fold-differences in PCR amplification. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression was measured to normalize the expression of each gene. All primers were synthesized at Invitrogen’s (Beijing, China) core facility. The primer sequences used (5′-3′) were as follows: PTEN sense: GGACGAACTGGTGTAATG, antisense: GCCTCTGACTGGGAATAG; PDCD4 sense: AGGCTGAGGCAGGAGAAT, antisense: TCCCACCAGTAATGACAAAA; GAPDH sense: GAAGGTGAAGGTCGGAGT, antisense: GAGATGGTGATGGGATTTC.
Western blot analysis
Total protein (50 μg) was separated by 10 or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto polyvinylidene fluoride membranes. Specific primary antibodies including rabbit monoclonal anti-PTEN, mouse monoclonal anti-Beclin-1 (1:500; Abcam), anti-Bcl-2, anti-Bax, anti-PDCD4, anti-p-AKT, anti-LC3B (rabbit polyclonal; 1:500, GeneTex), anti-caspase-3 (mouse polyclonal, 1:1000; Cell Signaling Technology), and horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (1:4000) were used for western blotting. Protein bands were visualized using a Super-Signal West Pico Kit (Thermo Scientific, Rockford, IL, USA) and ChemiDoc MP Imaging System with Image Lab™ Touch Software (Bio-Rad Laboratories, Inc, Hercules, CA, USA). An anti-GAPDH antibody (mouse monoclonal, 1:1000; Santa Cruz Biotechnology) was used as a standard for quantitative protein analysis.
Statistical analysis
Statistical significance was determined by an independent t-test for qRT-PCR, autophagy, and migration tests and by a paired t-test and one-way analysis of variance (ANOVA) for apoptosis and western blot analysis using GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA, USA). Mean differences were considered statistically significant when p < 0.05. Data are shown as the mean ± standard error of the mean (S.E.M.).
Results
Differential expression profile of microRNAs in keloid dermis
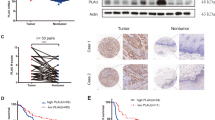
The microRNA microarray detected 14 upregulated and 15 downregulated miRs (p < 0.01; fold-change < 0.5 or >2) in the keloid dermis compared with expression in the normal dermis (Fig. 1a, Table 1). MiR-21-5p, miR-214-5p, miR-542-5p, miR-493-5p, and miR-409-3p expression was significantly upregulated in the keloid dermis (p = 0.0002, p = 0.04, p = 0.0499, p = 0.01, and p = 0.003, respectively; Fig. 1b) compared with expression in the normal dermis based on qRT-PCR.
Differential expression of microRNAs (miR) in the dermis derived from keloid and normal skin. a MiR expression profiles were detected by Agilent’s human microRNA microarray. Red dots in the volcano plot represent upregulated and downregulated miRs (n = 3; p < 0.05; fold-change < 0.5 or >2) in the keloid versus expression in the normal dermis. b Six differentially expressed miRs were verified by SYBR Green quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The expression of each miR was normalized to the expression of U6. Statistical significance was determined using an independent sample t-test. Data are shown as the means ± standard error of the mean (S.E.M; n = 8; ***p < 0.001, **p < 0.01, and *p < 0.05)
LC3B expression and migration are enhanced in keloid dermis fibroblasts and in vitro
Immunohistochemical analysis showed higher expression of the autophagy-related markers LC3B and Beclin-1 in fibroblasts of the margin dermis of the keloid compared with that in the adjacent normal skin (Fig. 2a–c). In addition, protein levels of LC3B-II were significantly upregulated in the perilesional tissue of the keloid compared with those in the extralesional skin based on western blot analysis (p = 0.04; Fig. 2d, e). These data suggested that autophagy is enhanced in the margin area of the keloid. These results were in accordance with clinical observations that the perilesional areas of the keloid display aggressive growth and invasion into normal skin.
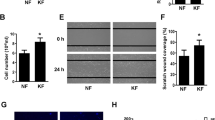
Expression of autophagy-related genes and migration capacity of fibroblasts in keloids and adjacent normal skin samples. a Specific keloid disease lesional sites consisting of extralesional (green: normal adjacent skin), perilesional (red: keloid margin), and intralesional (blue: keloid center) sites. b Image of LC3B and Beclin-1 expression based on immunohistochemical analysis in extralesional, perilesional, and intralesional sites of keloid disease based on photomerge software (black bar = 100 μm). c Red-boxed inserts show the positive expression of LC3B and Beclin-1 in fibroblasts of the keloid margin dermis and green-boxed inserts show the expression of LC3B and Beclin-1 in the normal adjacent skin dermis (white bar = 100 μm). Protein expression levels of LC3B and Beclin-1 in keloid tissue and adjacent normal skin samples (d, e; n = 4) and in keloid fibroblasts and those from adjacent normal skin (h, i; n = 3) based on western blot analysis (*p < 0.05). Western blot images are shown in d and h. Comparisons of cell migration capacities between keloid fibroblasts (KFs) and adjacent normal fibroblasts (NFs). f Representative micrographs of KFs and NFs subjected to in vitro scratch–wound migration assays at 0 and 48 h (bar = 100 μm). g The scratch areas filled by fibroblasts were observed at 48 h after scratching (0 h). The percentage of wound healed area of fibroblasts was compared between keloids and normal skin groups (n = 3, ***p < 0.001). Data are shown as means ± S.E.M
Scratch–wound healing migration assays showed that the percentage of the wound healed area of keloid fibroblasts (43.56 ± 3.17%) was significantly increased compared with that of adjacent normal skin fibroblasts (21.25 ± 3.28%; p < 0.0001; Fig. 2f, g). Protein levels of LC3B-II were also significantly increased in keloid fibroblasts (2.37 ± 0.32) compared with those in adjacent normal skin fibroblasts (0.93 ± 0.07) based on western blotting (p = 0.01; Fig. 2h, i).
The optimal doses of EB irradiation reduce autophagy and related gene expression in keloid fibroblasts
To investigate the optimal therapeutic dose of EB irradiation, apoptosis was measured in keloid fibroblasts following exposure to different doses (0, 5, 10, 15, 20, and 30 Gy) of EB irradiation. The percentage of apoptotic cells peaked at 20 Gy (57.78 ± 8.59%; untreated cells, 36.21% ± 2.43%), with a significant difference observed between 0 and 20 Gy groups (p < 0.05; Fig. 3a, b). The results indicated that 20-Gy EB irradiation has better therapeutic effects on keloids by inducing higher levels of apoptosis in keloid fibroblasts.
Electron beam (EB) irradiation at 20 Gy suppresses autophagy and the expression of autophagy-related genes in keloid fibroblasts. a, b Fibroblasts were exposed to single doses of 0, 5, 10, 15, 20, and 30 Gy of EB irradiation and cultured for 3 days. Then, apoptotic cells were detected using an Annexin V PE Apoptosis Detection Kit. The percentage of apoptotic cells peaked at 20 Gy. c One hour after a single 20-Gy dose of EB irradiation, autophagosomes with two membranes (black arrowheads) and autolysosomes with a single membrane (white arrowheads) were observed in cells by electron microscopy. d The autophagy-related genes LC3B-II and Beclin-1 were significantly downregulated compared with expression in controls based on western blotting. Western blot images are shown in e. Data are shown as means ± S.E.M (n = 3; *p < 0.05, **p < 0.01)
After exposure to 20 Gy of EB irradiation, fewer double-membrane autophagosomes and single-membrane autolysosomes were observed in keloid fibroblasts than in untreated cells by electron microscopy (Fig. 3c). Western blot analysis showed that the protein levels of autophagy-related genes, LC3B-II and Beclin-1, were also significantly downregulated (p = 0.04 and p = 0.003, respectively; Fig. 3d, e) upon 20 Gy of EB irradiation, compared with control levels. These results indicated that apoptosis was enhanced and that autophagy was inhibited after keloid fibroblasts were exposed to an optimal dose of therapeutic EB irradiation.
EB irradiation regulates the expression of miR-21-5p and apoptosis-associated genes in keloid fibroblasts
MiR-21-5p was found to be significantly downregulated after keloid fibroblasts were exposed to 20 Gy of EB irradiation, based on qRT-PCR, as compared with expression in untreated cells (p = 0.04; Fig. 4a). The mRNA and protein levels of PTEN and programmed cell death 4 (PDCD4) were significantly increased following miR-21-5p inhibition as determined by qRT-PCR (p = 0.03 and p = 0.007, respectively; Fig. 4b, c) and western blotting (p = 0.01 and p = 0.03, respectively; Fig. 4d, e); levels of p-AKT decreased in parallel (p = 0.04; Fig. 4d, e). In contrast, expression levels of the apoptosis-associated genes Bcl-2, Bax, and caspase-3 were significantly increased (p = 0.03, p = 0.01, and p = 0.04, respectively, Fig. 4d, f).
Electron beam (EB) irradiation regulates the expression of miR-21-5p and apoptosis-associated genes. a MiR-21-5p expression was found to be significantly downregulated after keloid fibroblasts were exposed to 20 Gy of EB irradiation, as compared with that in untreated cells, based on the TaqMan MicroRNA qRT-PCR Assay System. The mRNA expression of PTEN (b) and PDCD4 (c) was found to be significantly enhanced based on qRT-PCR. Western blot images are shown in d; the protein levels of PTEN, PDCD4 (e), and apoptosis-associated markers Bcl-2, Bax, and caspase-3 (f) were significantly increased based on western blotting. In contrast, the protein levels of p-AKT (e) were decreased. Data are shown as the means ± S.E.M (n = 3; *p < 0.05, **p < 0.01)
An miR-21-5p inhibitor regulates the expression of miR-21-5p and attenuates autophagy in keloid fibroblasts
The expression of miR-21-5p was significantly decreased in keloid fibroblasts transfected with the miR-21-5p inhibitor (25 nM) versus that with the corresponding inhibitor negative controls (p = 0.007; Fig. 5a). Cyto-ID Green autophagy detection by flow cytometry showed that the percentage of autophagy-positive cells was significantly decreased after miR-21-5p inhibitor transfection (43.4 ± 2.57%) compared with that in cells transfected with the corresponding inhibitor negative control (62.87 ± 2.67%; p = 0.002; Fig. 5b). Microscopic Cyto-ID Green fluorescence autophagy detection showed fewer autophagy-positive cells after miR-21-5p inhibitor transfection (25 nM) as compared with control levels (Fig. 5c). However, no significant differences in autophagy-positive cells were found between adjacent normal skin fibroblasts transfected with the miR-21-5p inhibitor (25 nM) and the negative control group using the Cyto-ID Green Autophagy Detection Kit by flow cytometry and fluorescence microscopy-based two-color detection (p = 0.29; Supplemental Fig. 1b, c).
The inhibition of miR-21-5p diminishes autophagy in keloid fibroblasts. a After the miR-21-5p inhibitor (25 nM) and corresponding inhibitor negative control were transfected into keloid fibroblasts for 48 h, the expression of miR-21-5p decreased significantly in cells transfected with the miR-21-5p inhibitor versus that in the corresponding inhibitor negative control groups, as determined using the TaqMan MicroRNA qRT-PCR Assay System. Autophagic cells were detected using the Cyto-ID Green Autophagy Detection Kit by flow cytometry and fluorescence microscopy-based two-color detection, respectively. b The percentage of autophagy-positive fibroblasts was significantly decreased among cells transfected with the miR-21-5p inhibitor compared with that in the corresponding inhibitor negative control group, based on flow cytometry. Data are shown as the means ± S.E.M (n = 3; **p < 0.01). c The number of autophagy-positive (green) fibroblasts decreased among cells transfected with the miR-21-5p inhibitor compared with that in corresponding inhibitor negative control or positive control groups, as determined by fluorescence microscopy. Scale bar: 100 μm. Positive control group: cells induced with rapamycin (500 nM) combined with chloroquine (10 μM); blank control group: cells cultured in DMEM supplemented with 10% FBS without induction or transfection
An miR-21-5p inhibitor regulates migration and autophagy-associated gene expression in keloid fibroblasts via AKT signaling and PTEN targeting
Scratch–wound healing migration assays were then performed after keloid fibroblasts were transfected with an miR-21-5p inhibitor and siPTEN. Results showed that the percentage of the wound healed area was significantly decreased in keloid fibroblasts (71.15 ± 1.38%) transfected with the miR-21-5p inhibitor (51.94 ± 2.07%; p < 0.001) and increased in cells transfected with siPTEN (83.69 ± 1.13%; p < 0.001). Furthermore, wound healing was completely recovered (p < 0.001) after the transfection of both the inhibitor and siPTEN (74.9 ± 1.4%; p < 0.001; Fig. 6a, b).
MiR-21-5p regulates the migration of keloid fibroblasts and the autophagy-associated marker LC3B via AKT signaling by targeting PTEN. Keloid fibroblasts were transfected with the miR-21-5p inhibitor (I) (25 nM) or the negative inhibitor control (IC) for 48 h, or were first transfected with a selected RNA duplex specific for PTEN mRNA (siPTEN; 60 nM) for 24 h and then transfected with the miR-21-5p inhibitor (I) (25 nM) or negative inhibitor control (IC) for 48 h. a Representative micrographs of fibroblasts in each transfection group subjected to in vitro scratch–wound healing migration assays at 0 and 48 h. b The percentages of wound healed area of fibroblasts were compared among each transfection group. Western blot images are shown in c. The expression levels of LC3B, Beclin-1 (d), PTEN, and p-AKT (e) were examined by western blot analysis. Levels of proteins were normalized to the expression of GAPDH. The western blot data are expressed as fold-change relative to the mean value of the control group. Statistical significance was obtained based on one-way ANOVA analysis. Data are shown as the mean ± S.E.M (n = 3; *p < 0.05, **p < 0.01, ***p < 0.001)
The expression of PTEN, p-AKT, and autophagy-related genes was also examined after keloid fibroblasts were transfected with an miR-21-5p inhibitor and siPTEN (Fig. 6c–e). The expression of PTEN was significantly increased in cells transfected with the miR-21-5p inhibitor (p < 0.001) and decreased among cells transfected with siPTEN (p < 0.01). In the group of keloid fibroblasts transfected with the miR-21-5p inhibitor, p-AKT expression was significantly decreased (p < 0.001) with a concomitant reduction in LC3B-II levels (p < 0.05). Furthermore, when the expression of PTEN was significantly decreased in the inhibitor and siPTEN group (p < 0.01), p-AKT expression was completely recovered (p < 0.001), whereas LC3B-II levels were partially rescued. Moreover, in the PTEN-knockdown only group, there was a remarkable increase in the expression of LC3B-II (p < 0.001) in keloid fibroblasts compared with those in the inhibitor plus siPTEN group. However, Beclin-1 expression was not significantly changed in each group (p > 0.05).
These data indicated that miR-21-5p downregulation inhibits migration and expression of the autophagy-associated marker LC3B in keloid fibroblasts via AKT signaling and PTEN targeting.
Nevertheless, the percentage of the wound healed area was not significantly decreased in normal skin fibroblasts transfected with the miR-21-5p inhibitor and was not increased in cells transfected with siPTEN (p > 0.05; Supplemental Fig. 2a, b), and the expression of PTEN, p-AKT, LC3B-II, and Beclin-1 was not significantly changed in each group (Supplemental Fig. 2c–e; p > 0.05).
An miR-21-5p inhibitor enhances apoptosis and regulates apoptosis-associated gene expression in keloid fibroblasts
The percentage of apoptotic cells was significantly increased in keloid fibroblasts transfected with the miR-21-5p inhibitor (25 nM; 55.15 ± 1.11%) versus that in the negative control group (40.97 ± 1.98%; p < 0.01; Fig. 7a).
The miR-21-5p inhibitor regulates apoptosis and apoptosis-associated genes in keloid fibroblasts. The miR-21-5p inhibitor (25 nM) and corresponding inhibitor negative control were transfected into separate groups of keloid fibroblasts for 48 h. a The percentage of apoptotic cells was significantly increased in keloid fibroblasts transfected with the miR-21-5p inhibitor at a final concentration of 25 nM versus that in the negative control group. b The mRNA expression of PTEN and PDCD4 was detected by qRT-PCR. Western blot images are shown in c. d The protein levels of PDCD4 and apoptosis-associated genes (Bcl-2, Bax, and caspase-3) were significantly increased in cells transfected with the miR-21-5p inhibitor versus levels in control cells. Data are shown as the means ± S.E.M. (n = 3; *p < 0.05, **p < 0.01)
Meanwhile, PTEN mRNA levels significantly increased in keloid fibroblasts transfected with the miR-21-5p inhibitor as determined by qRT-PCR (p = 0.01, Fig. 7b). Although no significant differences in PDCD4 mRNA expression were found between the two groups (p = 0.67; Fig. 7b), protein levels were upregulated (p = 0.02; Fig. 7c, d), whereas expression of the apoptosis-associated genes Bcl-2, Bax, and caspase-3 increased significantly (p = 0.04, p = 0.02, and p = 0.03, respectively; Fig. 7c, d).
The percentage of apoptotic cells and the protein expression levels of the apoptosis-associated genes Bcl-2, Bax, and caspase-3 did not significantly increase in adjacent normal skin fibroblasts transfected with the miR-21-5p inhibitor (25 nM) compared with that in the negative control group (p = 0.95; Supplemental Fig. 3a–c).
Discussion
The high level of keloid invasion and recurrence remains a challenge for clinical therapy due to unknown key pathogenic mechanisms. Cell migration is essential for wound healing and autophagy has been shown to modulate migration and metastasis [19, 20]; moreover, local migration and invasion into surrounding tissue represents the first step of metastasis. The autophagy-related marker LC3 is significantly downregulated in hypertrophic scars without aggressive growth [21]. The overexpression of LC3B both in the perilesion of keloid tissues and in accompanying fibroblasts, which have enhanced migration ability, suggested that autophagy is related to the local invasion of keloids; however, the regulatory mechanism needed to be investigated.
Autophagy and apoptosis often occur in the same cell to determine cell fate [22]. Enhanced antiapoptotic ability in fibroblasts is the important pathogenic mechanism underlying keloid recurrence and invasion [23]. Previously, EB irradiation was shown to induce apoptosis and enhance the expression of proapoptotic genes in keloid fibroblasts [8]. However, there have been no reports on autophagic pathways after the EB irradiation of keloids. During tumor progression, autophagy serves as a survival mechanism to help cells defend against the toxic effects of chemotherapy or radiation, whereas the inhibition of cytoprotective functions by autophagy enhances the radiosensitivity of tumor cells [24, 25]. Therefore, our results suggested that optimal therapeutic EB irradiation plays a role in preventing the recurrence of keloids by inhibiting cytoprotective autophagy, leading to keloid fibroblast apoptosis.
The Beclin-1 class III phosphatidylinositol-3-kinase complex drives autophagy activation. Moreover, formation of the LC3-conjugation system is responsible for vesicle elongation during autophagy [26]. Our results suggested that EB irradiation suppresses autophagy by downregulating the autophagy-related inducer Beclin-1 and preventing the formation of LC3B-II. However, the interplay between apoptosis and autophagy is complex. Accordingly, the interaction between apoptosis- and autophagy-associated genes plays an important role in crosstalk between these pathways. For example, autophagy has been shown to be inhibited by the binding of Bcl-2 to Beclin-1 or cleavage of the Beclin-1 complex by caspase-3 [22]. Our results suggested that EB irradiation induces apoptosis in keloid fibroblasts by upregulating the expression of proapoptotic Bax and caspase-3. Furthermore, the overexpression of Bcl-2 and caspase-3 might also play a role in diminishing autophagy by inhibiting Beclin-1; however, the regulatory mechanism underlying the interaction between apoptosis- and autophagy-associated genes needs to be investigated.
MiR-21 has been shown to promote radioresistance in various cancers by regulating apoptosis and autophagy [17]. Further, PTEN and PDCD4 are miR-21-target genes related to radiosensitivity. PTEN plays a critical role as a negative regulator of AKT signaling and miR-21 inhibition has been shown to increase cancer cell radiosensitivity by activating PTEN [27, 28]. However, it was previously unclear whether miR-21-5p expression alters or mediates keloid fibroblast autophagy after EB irradiation. Our findings revealed that miR-21-5p is overexpressed in the keloid dermis and that EB irradiation inhibits its expression, upregulating the expression of its target genes.
MiR-21 was found to promote the migration and invasion of cancers by regulating autophagy [29]. To determine the effects of miR-21-5p on keloid fibroblast autophagy, migration, and apoptosis and clarify the underlying molecular mechanism(s), an inhibitor was used to downregulate its expression. Our results showed that upon treatment with the miR-21-5p inhibitor, the tendency of keloid fibroblasts to undergo autophagy and apoptosis was comparable to that observed in EB-irradiated fibroblasts. Furthermore, the observed changes in miR-21-target gene expression and apoptosis- and autophagy-associated genes in fibroblasts after miR-21-5p inhibition were consistent with those observed in fibroblasts subjected to EB irradiation. Meanwhile, migration was inhibited, which was accompanied by the downregulation of LC3B-II, after keloid fibroblasts were transfected with the miR-21-5p inhibitor, whereas PTEN downregulation was found to reverse changes in migration and the expression of LC3B-II and p-AKT. LC3B participates in the regulation of tumor cell motility and its inhibition suppresses migration and invasion in various cancers [30,31,32]. Therefore, our results indicated that miR-21-5p plays a key role in autophagy and migration in keloid fibroblasts after EB irradiation by modulating PTEN and the AKT signaling pathway. However, organ cultures of keloid tissue have been reported to provide a quantitative, clinically relevant model to study keloid pathobiology and response to treatment [33, 34]. In future studies, using the keloid tissue organ culture system could support our current findings based on cells grown under two-dimensional cell culture condition.
In conclusion, our results indicated that EB irradiation can inhibit autophagy and increase apoptosis in keloid fibroblasts by reducing miR-21-5p expression, which was found to regulate migration and autophagy-associated gene expression via PTEN and p-AKT signaling. These data provide a possible mechanism wherein miR-21-5p regulates autophagy, migration, and apoptosis in EB-irradiated keloid fibroblasts, thereby preventing their local invasion and recurrence. Therefore, miR-21-5p could potentially be a new therapeutic target that regulates autophagy and migration, and this approach could be used instead of EB irradiation to control keloid relapse.
References
Shih B, Garside E, McGrouther DA, Bayat A. Molecular dissection of abnormal wound healing processes resulting in keloid disease. Wound Repair Regen. 2010;18:139–53.
Wolfram D, Tzankov A, Pülzl P, Piza-Katzer H. Hypertrophic scars and keloids–a review of their pathophysiology, risk factors, and therapeutic management. Dermatol Surg. 2009;35:171–81.
Syed F, Sanganee HJ, Singh S, Bahl A, Bayat A. Potent dual inhibitors of TORC1 and TORC2 complexes (KU-0063794 and KU-0068650) demonstrate in vitro and ex vivo anti-keloid scar activity. J Invest Dermatol. 2013;133:1340–50.
Luo L, Li J, Liu H, Jian X, Zou Q, Zhao Q, et al. Adiponectin is involved in connective tissue growth factor-induced proliferation, migration and overproduction of the extracellular matrix in keloid fibroblasts. Int J Mol Sci. 2017;18:E1044.
Jumper N, Hodgkinson T, Paus R, Bayat A. A Role for Neuregulin-1 in promoting keloid fibroblast migration via ErbB2-mediated signaling. Acta Derm Venereol. 2017;97:675–84.
Wang LZ, Ding JP, Yang MY, Chen B. Forty-five cases of chest keloids treated with subcutaneous super-tension-reduction suture combined with postoperative electron-beam irradiation. Dermatol Surg. 2014;40:1378–84.
Shen J, Lian X, Sun Y, Wang X, Hu K, Hou X, et al. Hypofractionated electron-beam radiation therapy for keloids: retrospective study of 568 cases with 834 lesions. J Radiat Res. 2015;56:811–7.
Tosa M, Ghazizadeh M, Shimizu H, Hirai T, Hyakusoku H, Kawanami O. Global gene expression analysis of keloid fibroblasts in response to electron beam irradiation reveals the involvement of interleukin-6 pathway. J Invest Dermatol. 2005;124:704–13.
Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res. 2012;18:370–9.
Yu T, Zuber J, Li J. Targeting autophagy in skin diseases. J Mol Med. 2015;93:31–8.
Ramkumar A, Murthy D, Raja DA, Singh A, Krishnan A, Khanna S, et al. Classical autophagy proteins LC3B and ATG4B facilitate melanosome movement on cytoskeletal tracks. Autophagy. 2017;13:1331–47.
Sharifi MN, Mowers EE, Drake LE, Collier C, Chen H, Zamora M, et al. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of paxillin with LC3. Cell Rep. 2016;15:1660–72.
Okuno R, Ito Y, Eid N, Otsuki Y, Kondo Y, Ueda K. Upregulation of autophagy and glycolysis markers in keloid hypoxic-zone fibroblasts: morphological characteristics and implications. Histol Histopathol. 2018;33:1075–87.
Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–13.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60.
Duffield JS, Grafals M, Portilla D. MicroRNAs are potential therapeutic targets in fibrosing kidney disease: lessons from animal models. Drug Discov Today Dis Models. 2013;10:e127–35.
Liu J, Zhu H, Yang X, Ge Y, Zhang C, Qin Q, et al. MicroRNA-21 is a novel promising target in cancer radiation therapy. Tumour Biol. 2014;35:3975–9.
Liu Y, Wang X, Yang D, Xiao Z, Chen X. MicroRNA-21 affects proliferation and apoptosis by regulating expression of PTEN in human keloid fibroblasts. Plast Reconstr Surg. 2014;134:561e–73e.
Tuloup-Minguez V, Hamaï A, Greffard A, Nicolas V, Codogno P, Botti J. Autophagy modulates cell migration and β1 integrin membrane recycling. Cell Cycle. 2013;12:3317–28.
Dower CM, Wills CA, Frisch SM, Wang HG. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14:1110–28.
Shi JH, Hu DH, Zhang ZF, Bai XZ, Wang HT, Zhu XX, et al. Reduced expression of microtubule-associated protein 1 light chain 3 in hypertrophic scars. Arch Dermatol Res. 2012;304:209–15.
Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94.
Lu F, Gao J, Ogawa R, Hyakusoku H, Ou C. Fas-mediated apoptotic signal transduction in keloid and hypertrophic scar. Plast Reconstr Surg. 2007;119:1714–21.
Yang Y, Yang Y, Yang X, Zhu H, Guo Q, Chen X, et al. Autophagy and its function in radiosensitivity. Tumour Biol. 2015;36:4079–87.
Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48.
Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–90.
Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus. 2013;26:823–31.
Wang W, Zhao J, Wang H, Sun Y, Peng Z, Zhou G, et al. Programmed cell death 4 (PDCD4) mediates the sensitivity of gastric cancer cells to TRAILinduced apoptosis by down-regulation of FLIP expression. Exp Cell Res. 2010;316:2456–64.
Li S, Zeng X, Ma R, Wang L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018;16:2038–45.
Hamurcu Z, Delibaşı N, Geçene S, Şener EF, Dönmez-Altuntaş H, Özkul Y, et al. Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin β1/ Src signaling in triple negative breast cancer cells. J Cancer Res Clin Oncol. 2018;144:415–30.
Mowers EE, Sharifi MN, Macleod KF. Novel insights into how autophagy regulates tumor cell motility. Autophagy. 2016;12:1679–80.
Tang Z, Zhang N, Di W, Li W. Inhibition of microtubule-associated protein 1 light chain 3B via small-interfering RNA or 3-methyladenine impairs hypoxia-induced HO8910PM and HO8910 epithelial ovarian cancer cell migration and invasion and is associated with RhoA and alterations of the actin cytoskeleton. Oncol Rep. 2015;33:1411–17.
Bagabir R, Syed F, Paus R, Bayat A. Long-term organ culture of keloid disease tissue. Exp Dermatol. 2012;21:376–81.
Syed F, Bagabir RA, Paus R, Bayat A. Ex vivo evaluation of antifibrotic compounds in skin scarring: EGCG and silencing of PAI-1 independently inhibit growth and induce keloid shrinkage. Lab Invest. 2013;93:946–60.
Acknowledgements
This work was supported by CAMS Innovation Fund for Medical Sciences (Grant no. 2016-I2M-1-017), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT32015), and the National Natural Science Foundation of China (Grant No. 81171817).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yan, L., Wang, LZ., Xiao, R. et al. Inhibition of microRNA-21-5p reduces keloid fibroblast autophagy and migration by targeting PTEN after electron beam irradiation. Lab Invest 100, 387–399 (2020). https://doi.org/10.1038/s41374-019-0323-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0323-9
This article is cited by
-
MicroRNAs Associated with Keloids Identified by Microarray Analysis and In Vitro Experiments
Molecular Biotechnology (2024)
-
Characterization of the skin keloid microenvironment
Cell Communication and Signaling (2023)
-
LncRNA H19 promotes keloid formation through targeting the miR-769-5p/EIF3A pathway
Molecular and Cellular Biochemistry (2021)
-
Epigenetic modification mechanisms involved in keloid: current status and prospect
Clinical Epigenetics (2020)
-
Flaming the fight against cancer cells: the role of microRNA-93
Cancer Cell International (2020)