Abstract
The exact role of autophagy in breast cancers remains elusive. In this study, we explored the potential functions of autophagy-related 7 (Atg7) in breast cancer cell lines and tissues. Compared to normal breast tissue, a significantly lower expression of Atg7 was observed in triple-negative breast cancer (TNBC), but not other subtypes. A higher Atg7 expression was significantly associated with favorable clinicopathologic factors and better prognostic outcomes in patients with TNBC. Reflecting the clinical and pathologic observations, Atg7 was found to inhibit proliferation and migration, but promotes apoptosis in TNBC cell lines. Furthermore, Atg7 suppressed epithelial–mesenchymal transition through inhibiting aerobic glycolysis metabolism of TNBC cells. These findings provided novel molecular and clinical evidence of Atg7 in modulating the biological behavior of TNBC, thus warranting further investigation.
Similar content being viewed by others
Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death among women worldwide [1]. Triple-negative breast cancer (TNBC), characterized by lack of estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (HER-2), accounts for 15–20% of breast cancers [2]. Cytotoxic chemotherapy remains the mainstay for the treatment of TNBC due to lack of targeted therapies. Moreover, TNBC is prone to recurrence after treatment and thus is associated with a worse prognosis [3]. Therefore, it is necessary to identify novel biomarkers with prognostic significance and therapeutic implications in the pursuit of precision medicine.
As a conserved evolutionary process, autophagy is the only mechanism to degrade large structures such as organelles and proteins for cell to survive under stress [4]. Alteration of autophagy occurs in multiple cancer types, including lung, colorectal, and breast primaries [5,6,7]. Recent studies in this regard have indicated a paradoxical role of autophagy in cancer progression: it prevents initiation of some cancer types, but supports tumor growth in others [8]. Thus, the importance of autophagy-related genes in tumorigenesis and treatment have become increasingly recognized. Research to that end has demonstrated that downregulation of Atg5-dependent autophagy promotes breast cancer metastasis [9], while LC3 and Beclin-1 are required for cell proliferation, survival, migration, and invasion, and may contribute to tumor progression and metastasis in TNBC. Moreover, LC3B has been shown to be a poor prognostic indicator‘ in TNBC [10].
Autophagy-related 7 (Atg7), the product of autophagy-related gene 7 and also known as ubiquitin E1-like activating enzyme, is essential for the assembly and expansion of autophagosomal membranes [4]. Atg7-deficient mice have demonstrated impairment of both constitutive and starvation-induced autophagy and died soon after birth due to respiratory distress [11]. Recent studies have shown that Atg7 is overexpressed in breast cancer cells, and Atg7 rs8154 (A > G) polymorphism is significantly associated with breast cancer-specific survival [12]. Furthermore, silencing Atg7 resulted in significantly impaired high-LC3B triple-negative tumor growth in vivo [13]. In addition, Atg7 silencing in MCF-7 breast cancer cells led to enhanced efficacy and apoptotic effect of docetaxel and exhibited improved anticancer effects [14, 15]. These observations have strongly suggested a critical role of Atg7 in breast cancer progression.
In this study, we examined Atg7 expression in TNBC tissues to further assess its prognostic value. We also explored the influence of Atg7 on apoptosis, proliferation, migration, epithelial–mesenchymal transition (EMT), and energy metabolism in breast cancer cells. Our findings suggested that Atg7 might serve as a favorable prognostic indicator for TNBC. The molecule exerts its effects by inhibiting proliferation and migration, and promoting apoptosis of breast cancer cells. In addition, Atg7 downregulates aerobic glycolysis metabolism, thus inhibiting EMT.
Materials and methods
Patients and tissue specimens
This study was approved by the human ethics review committee of China Medical University (Shenyang, China). Breast cancer tissues from 90 patients and benign breast tissues from 9 patients with mastopathy (from 2014 to 2016) were obtained from the First Affiliated Hospital of China Medical University and used for immunohistochemical staining. Breast cancer subtypes were characterized as follows: Luminal A, Luminal B, HER-2, and TNBC.
Tissue microarrays of 164 TNBC were purchased from Outdo Biotech (Shanghai, China), along with the patients’ clinicopathologic characteristics, including the patient’s age, histologic grading, tumor size, Ki-67 proliferation index, lymphovascular invasion, and clinical (tumor node metastasis (TNM)) stage.
In addition, a separate cohort of 42 patients with TNBC from 2009 to 2013 were also included. All 42 patients were diagnosed as having invasive ductal carcinoma and underwent breast conservation surgery. The median follow-up of these patients was 68 months.
Immunohistochemical staining
The expressions of Atg7 was detected by immunohistochemical staining using a two-step protocol according to the manufacturer’s instructions (MXB, KIT-9710, China). The paraffin blocks were cut at 4 μm thickness. The sections were incubated overnight at 4 °C with anti-Atg7 (Sigma; HPA007639; dilution 1:100), followed by incubation at room temperature with biotinylated secondary antibody for 10 min. The negative control was established with the primary antibody replaced by phosphate-buffered saline (PBS). After DAB (3,3′-diaminobenzidine) staining, all sections were counter stained with hematoxylin before visualization.
The staining index (SI; range 0–12) was calculated by multiplying the staining intensity score on a scale of 0–3 by the proportion score on a scale of 1–4 (1, <25%; 2, 25–50%; 3, 50–75%; 4, >75%). SI ≥6 was defined as high expression and SI <6 was defined as low expression.
Cell culture and transfection
The cell lines were obtained from the cell bank of type culture collection of Chinese academy of sciences (Shanghai, China). MDA-MB-231 cells were cultured in L15 media (Sigma, USA); BT-549 cells were cultured in 1640 (Hyclone, USA), MCF-7, T47D, and SKBR-3 cells were cultured in DuIbecco’s modified EagIe’s medium (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (Clark, USA), 100U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. pcNDA 3.1 (Invitrogen, USA) was used as a cloning vector and control group. Lipofectamine 3000 (Invitrogen, USA) was used for transient transfection.
Western blot analysis
Cells were harvested and protein quantitation was conducted. Protein samples (40 μg) were separated on a 10% sodium dodecyl sulfate- polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes. Tris-buffered saline (containing 0.1% Tween-20 with 5% bovine serum albumin) was used as a blocking buffer. PVDF membranes were incubated with antibody raised against Atg7 (Sigma; A2856), Tubulin (Sigma; T6199), N-cadherin, SMA, Vimentin, E-cadherin, Slug, Snail (CST Kit, 9782T), cleaved caspase-3 (CST; 9664S), cleaved poly (ADP-ribose) polymerase (PARP) (CST; 5625S), or Bcl-2 (Santa Cruz, sc-509) overnight at 4 °C and washed for three times with Tris-buffered saline and Tween-20, followed by incubation with a horse radish peroxidase-linked secondary antibody. Blots were developed with ECL reagent (Tanon, China).
Virus production and infection
Atg7 knockdown was performed using short hairpin RNA (shRNA), and the sequence was generated by GeneChem Co., Ltd. (Shanghai, China). A scrambled shRNA was used as a negative control. The shRNA expression vectors were transfected into HEK293T cells using Lipofectamine 3000 reagent (Invitrogen). The supernatant fraction containing lentiviral particles was harvested at 24 and 48 h, respectively. MCF-7 cells were infected in the presence of 8 μg/ml polybrene (Santa Cruz) and then were treated with puromycin (Sigma) for 2 weeks.
Apoptosis assay
Cells were seeded in 6-well plates and were transfected for 36 h. Cells/tubes (5 × 105) were washed with PBS twice and were incubated with 200 μl binding buffer containing 5 μl of Annexin V-fluorescein isothiocyanate and propidium iodide each at room temperature for 15 min in the dark. Apoptosis was analyzed by flow cytometry (BD Flow Cytometer, USA). At least 3 × 104 events were collected for each sample at a flow rate of 80–100 cells/s.
Cell proliferation and migration assay
Following transfection with vector and pcDNA3.1-Atg7 for 24 h, cells were detached with trypsin and re-suspended as single-cell suspensions in low-serum L15. A total of 5 × 103 cells/well were added in 96-well plates. The cells were stained by CCK8 (Bimake, USA) after being planted for 24, 48, and 72 h, each group contained three parallel wells. Absorbance at 450 nm was measured to detect cell proliferation rate.
To examine cell migration, a total of 5 × 104 cells in low-serum L15 were added to the upper chamber (Corning, USA) and the lower chamber was filled with 0.6 ml of L15 supplemented with 10% FBS. The cells were allowed to migrate for 24 h and then were fixed with 4% anhydrous ethanol for 10 min at room temperature. The non-migrating cells on the upper side of the insert were removed with a cotton bar, while the migrated cells were stained by hematoxylin for 20 min at room temperature. The chambers were washed three times with PBS before imaging (Nikon DS-Fi2, Japan).
Measurement of OCR and ECAR
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XFp analyzer (Seahorse Bioscience, 103020-100). In brief, cells were plated on Seahorse XFp plates for 24 h at a concentration of 5 × 103 cells/well. Cells incubated at 37 °C for 12 h on probe plate and FCCP, oligomycin, and Rotenone/antimycin A were injected into the medium at final concentrations of 1 μM, 0.5 mM, and 1 mM, respectively. For ECAR, cells were plated on Seahorse XFp plates for 12–15 h at a concentration of 5 × 103 cells/well. Glucose, oligomycin, and 2-deoxyglucose were added to the medium at final concentrations of 10 mM, 1 μM, 50 mM, respectively. The OCR and ECAR were automatically recorded and calculated by the Seahorse XFp software as per the manufacture’s recommendation.
Statistical analysis
Nonparametric test were used to analyze the differences in the expression of Atg7 between breast cancer and adjacent benign breast tissues. Pearson's χ2 were used to analyze the correlation between Atg7 expression and clinicopathological parameters. Log-rank test of Kaplan–Meier method and multivariate Cox regression were conducted to explore the correlation between Atg7 expression and prognosis in TNBC patients. SPSS 19.0 was used for statistical analysis and P < 0.05 was considered statistically significant.
Results
Atg7 expression has prognostic value in TNBC
Immunohistochemical analysis of 90 breast cancer and 9 mastopathy tissues showed that Atg7 expression was significantly lower in TNBC when compared to mastopathy tissues (P < 0.001), while no significant difference was found between other breast cancer subtypes and mastopathy (Fig. 1a, c). Moreover, Atg7 expression in TNBC was significantly lower when compared to adjacent benign glandular tissues (P < 0.0001) (Fig. 1b, d).
Autophagy-related 7 (Atg7) expression is significantly decreased in triple-negative breast cancer (TNBC). a Representative weak, moderate, and strong Atg7 staining in breast cancer tissue (original magnification ×200). b Representative Atg7 staining in TNBC and adjacent non-tumoral tissue. c Atg7 expression in different subtypes of breast cancer and mastopathy tissues. d Atg7 expression in 10 TNBC and adjacent non-tumoral tissues (****P < 0.0001)
We next investigated the correlation of Atg7 expression and clinicopathologic parameters of TNBC. Atg7 expression was examined by immunohistochemical analysis of tissue microarrays consisting 164 TNBCs. As shown in Table 1, high Atg7 expression was more frequently seen in elderly patients, lower histological grade, lower Ki-67 proliferation index, negative lymph node status, and lower TNM stage. However, it was significantly associated with tumor size.
To further assess the prognostic significance of Atg7, single-factor log-rank test was performed using the cohort of 42 TNBC patients. To that end, a higher Atg7 expression was associated with a significantly favorable survival (Table 2 and Fig. 2). Multivariate Cox regression analysis further confirmed that Atg7 was an independent prognostic factor for OS in TNBC patients adjusted by age and TNM stage (adjusted hazard ratio (HR) = 0.302, 95% confidence interval (CI): 0.115–0.791, P = 0.015) (Table 2).
A higher autophagy-related 7 (Atg7) expression is associated with a favorable overall survival in patients with triple-negative breast cancer (TNBC). Notes: Overall survival of cohort of 42 patients with TNBC. Survival curves for ATG7-negative (red line) and ATG7-positive (blue line) patients (P = 0.005)
Atg7 promotes apoptosis of TNBC cells
We next examined the baseline expression of several major Atg proteins in multiple breast cancer cell lines. To that end, Atg7 expression in MDA-MB-231 and BT-549 cells (both TNBC) was much lower than that of MCF-7, T47D (both luminal), and SKBR-3 (HER-2+) cells. Besides, Atg5, P62, and LC3 indicated lower autophagic level in TNBC cells compared to none in TNBC cell lines (Fig. 3a). Given that Atg7 deficiency suppresses apoptosis in a mouse model [16], cell apoptosis assays were carried out after transfection of Atg7 into TNBC cells to further explore the potential mechanism of the molecule in cell death. Flow cytometry analyses indicated that overexpression of Atg7 increased the number of cells in early and middle apoptosis stages (Fig. 3b, c). Furthermore, western blot analyses showed that cleaved caspase-3 and cleaved PARP, the hallmarks of apoptosis, were upregulated, and the anti-apoptosis protein Bcl-2 was reduced by overexpressing Atg7 (Fig. 3d). These findings suggest that Atg7 promotes apoptosis of TNBC cells.
Autophagy-related 7 (Atg7) promotes apoptosis of triple-negative breast cancer (TNBC) cells. a Western blot analysis of Atg7, Atg5, Beclin-1, P62, and LC3 expression in multiple breast cancer cell lines. b, c Cells were stained with fluorescein isothiocyanate (FITC)-conjugated Annexin V and propidium iodide after transfection and analyzed by flow cytometry in MDA-MB-231 and BT-549 cells, respectively. d Western blot analysis of apoptosis-related proteins in TNBC cells transfected with and without Atg7
Atg7 inhibits migration and EMT in TNBC cells
Cell migration assay was next conducted using MDA-MB-231 and BT-549 cells transfected with pcDNA3.1-Atg7. As shown in Fig. 4a, b, there was significantly reduced cell migration in Atg7-transfected cells when compared to the control group. Further, Atg7 knockdown in MCF-7 cells using shRNA resulted in significantly induced cell migration, whereas restoration of Atg7 markedly dampened the cell migration ability (Fig. 4c, d).
Autophagy-related 7 (Atg7) inhibits migration and epithelial–mesenchymal transition (EMT) in triple-negative breast cancer (TNBC) cells. a, b MDA-MB-231 and BT-549 cells were transfected with vector and pcDNA3.1-Atg7, respectively, followed by migration assay as described in the Materials and methods. c Migration assays in Atg7-knockdown MCF-7 cells followed by restoration of Atg7. d Western blot analyses confirmed the successful knockdown and restoration of Atg7 in MCF-7 cells. e Overexpressing Atg7-induced alteration of EMT markers in TNBC by western blot analysis
EMT is believed to promote tumor initiation and progression [17,18,19]. Autophagy activation has been reported to reverse EMT progress through degradation of the EMT regulators SNAIL and TWIST in breast cancer models [20]. We thus asked whether Atg7 could affect EMT in TNBC cells. To that end, overexpression of Atg7 resulted in downregulation of N-cadherin, SMA, vimentin, Snail and Slug (known as “acquired markers” of EMT), and upregulation of E-cadherin (“attenuated marker” of EMT) (Fig. 4e). These observations suggest that Atg7 inhibits migration and EMT in TNBC cells.
Atg7 inhibits TNBC cell proliferation and prevents EMT through blocking aerobic glycolysis metabolism
To investigate whether Atg7 has impact on proliferation of TNBC cells, pcDNA3.1-Atg7 was transfected into MDA-MB-231 and BT-549 cells, followed by cell proliferation assays using CCK8 Kit. As shown in Fig. 5a, b, proliferation of TNBC cells was suppressed by Atg7. Moreover, Atg7-knockdown-induced MCF-7 cell proliferation was significantly inhibited by restoration of Atg7.
Autophagy-related 7 (Atg7) inhibits proliferation and mitochondria metabolism in triple-negative breast cancer (TNBC) cells. a, b MDA-MB-231 and BT-549 cells were transfected with vector and pcDNA3.1-Atg7, absorbance at 450 nm was measured after 24, 48, and 72 h. c MCF-7 cell proliferation after Atg7 knockdown and re-expression in knockdown cells. d, e The effect of Atg7 in oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of MDA-MB-231 and BT-549 cells. Data were shown as mean ± SEM of ≥3 technical replicates. Each experiment was repeated three times. f The effect of oligomycin on Atg7-induced inhibition of N-cadherin by western blot analysis
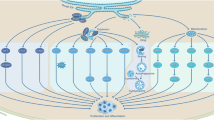
We have previously shown that Atg7 inhibits the Warburg effect, resulting in reduced EMT in HeLa and HEK293T cells [21]. We thus examined both OCR and ECAR of TNBC cells to further explore the potential roles of Atg7 in this regard. On OCR, the basal consumption rate and the spare respiratory capacity were both reduced by overexpressing Atg7. On ECAR, Atg7 led to decreased levels of lactate production in MDA-MB-231 and BT-549 cells. In addition, oligomycin (an aerobic glycolysis accelerator) not FCCP (an oxidative phosphorylation activator) recovered Atg7-induced loss of N-cadherin (Fig. 5f). Furthermore, we examined mitochondrial-specific proteins of mitofilin and VADC and found that Atg7 had no effect on the amount of mitochondria in cells (Fig. S1). These results suggest that Atg7 may inhibit EMT in TNBC cells through regulating aerobic glycolysis metabolism Fig. 6.
Model for autophagy-related 7 (Atg7) modulates tumor progression in triple-negative breast cancer. Atg7 leads to upregulated autophagy, apoptosis level, and regulate tumor proliferation and migration in TNBC cells. Atg7 inhibits aerobic glycolysis in the mitochondria, thereby inhibiting EMT. See text for details
Discussion
Given the widely recognized role of autophagy in tumorigenesis, there has been extensive research to identify novel autophagy-related treatment targets and prognostic markers. On the other hand, the role of Atg7, a key autophagy-related molecule, in tumor development remains inconclusive [22,23,24,25]. Until now, the exact role of Atg7 in breast carcinogenesis and prognosis is largely unclear. In this study, we have found that Atg7 plays a significant role in regulating proliferation, apoptosis, migration, EMT, and energy metabolism in TNBC cells. More importantly, our observations have suggested that Atg7 may serve as a prognostic marker in patients with TNBC.
There have been controversial data regarding Atg7 expression in breast cancer. An early study found that Atg7 was overexpressed in breast cancer cells [12]. In our study, however, a significantly lower level of Atg7 was observed in TNBC, but not other breast cancer subtypes, when compared to adjacent non-tumoral tissues. The variation in the levels of autophagy as represented by Atg7 among breast cancer subtypes suggests that it might play an important role in the development and progression of TNBC. Our findings of Atg7 as an independent prognostic factor in the cohort of 42 TNBC patients further supports this speculation.
The therapeutic effects of autophagy in TNBC remains debatable. Previous studies have found that in TNBC cells, autophagy induction may mediate anticancer effects of dihydroartemisinin, while cell-protective autophagy weakens the therapeutic effect of agents targeting insulin-like growth factor-1 receptor [26, 27]. However, the exact role of Atg7 on the initiation and progression of TNBC is largely unknown. In the current study, we found that Atg7 overexpression diminished proliferation and migration in MDA-MB-231 and BT-549, two TNBC cell lines. In addition, Atg7 knockdown in MCF-7 cells (non-TNBC) resulted in accelerated cell proliferation and migration, which were reversed by restoration of Atg7 in the knockdown cells. These observations have demonstrated a favorable function of Atg7 in suppressing TNBC progression.
Autophagy and apoptosis are two important cellular processes with complex and intersecting protein networks, both of which play major roles in determining cell fate, while some autophagy-related proteins are proved to possess pro- or anti-apoptosis functions [28,29,30,31]. Herein, we showed that Atg7 induced apoptotic markers cleaved PARP and cleaved caspase-3, and reduced anti-apoptotic protein Bcl-2 in TNBC cells. These findings account for the observation that Atg7 promotes apoptosis in TNBC cells, and further lead to the belief that the two cellular signaling pathways indeed interact and crosstalk in TNBC cells.
Atg7 was reported to enhance the expression of E-cadherin on chick embryo, and in HCT116 cells, a human colon cancer line [32]. Similarly, our study showed that Atg7 inhibited EMT progress as evidenced by the upregulation of E-cadherin (the so-called “attenuated marker” of EMT) and downregulation of N-cadherin, SMA, and Vimentin (known as “acquired markers” of EMT) in TNBC cells. Furthermore, we have previously shown that inhibition of the Warburg effect by Atg7 results in reduced EMT in HeLa and HEK293T cells. Our study demonstrated that Atg7 reduced mitochondrial metabolism by inhibiting both OCR and ECAR. What is more, oligomycin, an aerobic glycolysis accelerator, reversed Atg7-induced inhibition of EMT in TNBC cells. The collective findings thus far indicate that Atg7 prevents EMT mainly through inhibiting aerobic glycolysis in TNBC cells. Therefore, the molecule may provide a potential therapeutic target for this aggressive tumor type.
In summary, we have found that Atg7 inhibits proliferation and migration, and promotes apoptosis of TNBC cells. Furthermore, Atg7 prevents EMT through downregulation of cell aerobic metabolism. Moreover, our observations have suggested that Atg7 may be a potential prognostic marker and therapeutic target. The underlying regulatory mechanism of Atg7 on the biological behavior of TNBC cells requires further investigation given that cytotoxic chemotherapy remains the only systemic treatment modality for this highly aggressive subtype of breast cancer. Larger cohort studies are needed to confirm the prognostic significance of Atg7 in the pursuit of precision medicine.
References
Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM. Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. CA Cancer J Clin. 2017;67:378–97.
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–67.
Penault-Llorca F, Radosevic-Robin N. Biomarkers of residual disease after neoadjuvant therapy for breast cancer. Nat Rev Clin Oncol. 2016;13:487–503.
Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32.
Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–43.
Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell. 2016;166:288–98.
Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–42.
Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8.
Han Q, Deng Y, Chen S, Chen R, Yang M, Zhang Z, et al. Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci Rep. 2017;7:4759.
Hamurcu Z, Delibasi N, Gecene S, Sener EF, Donmez-Altuntas H, Ozkul Y, et al. Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin beta1/ Src signaling in triple negative breast cancer cells. J Cancer Res Clin Oncol. 2018;144:415–30.
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34.
Zhou J, Hang D, Jiang Y, Chen J, Han J, Zhou W, et al. Evaluation of genetic variants in autophagy pathway genes as prognostic biomarkers for breast cancer. Gene. 2017;627:549–55.
Lefort S, Joffre C, Kieffer Y, Givel AM, Bourachot B, Zago G, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10:2122–42.
Gong C, Hu C, Gu F, Xia Q, Yao C, Zhang L, et al. Co-delivery of autophagy inhibitor ATG7 siRNA and docetaxel for breast cancer treatment. J Control Rel. 2017;266:272–86.
Xue LY, Chiu SM, Oleinick NL. Atg7 deficiency increases resistance of MCF-7 human breast cancer cells to photodynamic therapy. Autophagy. 2010;6:248–55.
Kessel DH, Price M, Reiners JJ Jr. ATG7 deficiency suppresses apoptosis and cell death induced by lysosomal photodamage. Autophagy. 2012;8:1333–41.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial–mesenchymal transition and generation of breast cancer stem cells. Cancer Res. 2010;70:3005–8.
Ledford H. Cancer theory faces doubts. Nature. 2011;472:273.
Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial–mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238–50.
Feng Y, Liu J, Guo W, Guan Y, Xu H, Guo Q, et al. Atg7 inhibits Warburg effect by suppressing PKM2 phosphorylation resulting reduced epithelial–mesenchymal transition. Int J Biol Sci. 2018;14:775–83.
Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS ONE. 2011;6:e18691.
Desai S, Liu Z, Yao J, Patel N, Chen J, Wu Y, et al. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J Biol Chem. 2013;288:9165–76.
McCoy F, Hurwitz J, McTavish N, Paul I, Barnes C, O'Hagan B, et al. Obatoclax induces Atg7-dependent autophagy independent of beclin-1 and BAX/BAK. Cell Death Dis. 2010;1:e108.
Gong K, Chen C, Zhan Y, Chen Y, Huang Z, Li W. Autophagy-related gene 7 (ATG7) and reactive oxygen species/extracellular signal-regulated kinase regulate tetrandrine-induced autophagy in human hepatocellular carcinoma. J Biol Chem. 2012;287:35576–88.
Liu Q, Zhou X, Li C, Zhang X, Li CL. Rapamycin promotes the anticancer action of dihydroartemisinin in breast cancer MDA-MB-231 cells by regulating expression of Atg7 and DAPK. Oncol Lett. 2018;15:5781–6.
Wu W, Ma J, Shao N, Shi Y, Liu R, Li W, et al. Co-targeting IGF-1R and autophagy enhances the effects of cell growth suppression and apoptosis induced by the IGF-1R inhibitor NVP-AEW541 in triple-negative breast cancer cells. PLoS ONE. 2017;12:e0169229.
Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. et al. Autophagy and apoptosis: where do they meet? Apoptosis. 2014;19:555–66.
Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. et al. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900.
Shi M, Zhang T, Sun L, Luo Y, Liu DH, Xie ST, et al. Calpain, Atg5 and Bak play important roles in the crosstalk between apoptosis and autophagy induced by influx of extracellular calcium. Apoptosis. 2013;18:435–51.
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32.
Lu WH, Wang G, Li Y, Li S, Song XY, Wang XY, et al. Autophagy functions on EMT in gastrulation of avian embryo. Cell Cycle. 2014;13:2752–64.
Funding
This work was supported by National Key R&D Program of China (2016YFC1302400); Ministry of Education Innovation Team Development Plan (IRT_17R107); Natural Science Foundation of China (81770001, 31300963, 81130042, 31171323, 81502438); Natural Science Foundation of Liaoning Province of China (LFWK201725, LQNK201747).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, M., Liu, J., Li, S. et al. Autophagy-related 7 modulates tumor progression in triple-negative breast cancer. Lab Invest 99, 1266–1274 (2019). https://doi.org/10.1038/s41374-019-0249-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0249-2
This article is cited by
-
Insights on E1-like enzyme ATG7: functional regulation and relationships with aging-related diseases
Communications Biology (2024)
-
MAT as a promising therapeutic strategy against triple-negative breast cancer via inhibiting PI3K/AKT pathway
Scientific Reports (2023)
-
Autophagy regulation by RNA alternative splicing and implications in human diseases
Nature Communications (2022)
-
Hypoxia-autophagy axis induces VEGFA by peritoneal mesothelial cells to promote gastric cancer peritoneal metastasis through an integrin α5-fibronectin pathway
Journal of Experimental & Clinical Cancer Research (2020)