Abstract
Mesenchymal stem cells (MSCs) are multipotent progenitors that can differentiate into multiple lineages including osteoblastic lineage. Osteogenic differentiation of MSCs is a cascade that recapitulates most, if not all, of the molecular events occurring during embryonic skeletal development, which is regulated by numerous signaling pathways including bone morphogenetic proteins (BMPs). Through a comprehensive analysis of the osteogenic activity, we previously demonstrated that BMP9 is the most potent BMP for inducing bone formation from MSCs both in vitro and in vivo. However, as one of the least studied BMPs, the essential mediators of BMP9-induced osteogenic signaling remain elusive. Here we show that BMP9-induced osteogenic signaling in MSCs requires intact Notch signaling. While the expression of Notch receptors and ligands are readily detectable in MSCs, Notch inhibitor and dominant-negative Notch1 effectively inhibit BMP9-induced osteogenic differentiation in vitro and ectopic bone formation in vivo. Genetic disruption of Notch pathway severely impairs BMP9-induced osteogenic differentiation and ectopic bone formation from MSCs. Furthermore, while BMP9-induced expression of early-responsive genes is not affected by defective Notch signaling, BMP9 upregulates the expression of Notch receptors and ligands at the intermediate stage of osteogenic differentiation. Taken together, these results demonstrate that Notch signaling may play an essential role in coordinating BMP9-induced osteogenic differentiation of MSCs.
Similar content being viewed by others
Introduction
Mesenchymal stem cells (MSCs) are multipotent progenitors which can undergo self-renewal and differentiate into multi-lineages, such as osteogenic, chondrogenic, and adipogenic lineages [1, 2]. Osteogenic differentiation of MSCs is a cascade that recapitulates most, if not all, of the molecular events occurring during embryonic skeletal development [3]. Bone morphogenetic proteins (BMPs) play an important role during development [4,5,6] and have been shown to regulate stem cell proliferation and osteogenic differentiation [7,8,9]. BMPs belong to the TGFβ superfamily and consist of at least 14 members in humans [5, 6, 8, 10, 11].
Through a comprehensive analysis of the osteogenic activity, we previously found that BMP9 is one of the most potent BMPs among the 14 types of BMPs in inducing osteogenic differentiation of MSCs both in vitro and in vivo [5, 10,11,12,13]. BMP9 (also known as growth differentiation factor 2, or GDF-2) was identified in the developing mouse liver [14]. BMP9 has also been shown to play roles in inducing and maintaining the cholinergic phenotype of embryonic basal forebrain cholinergic neurons [15], inhibiting hepatic glucose production and inducing the expression of key enzymes of lipid metabolism [16], and regulating endothelial function and angiogenesis [17]. Through transcriptomic profiling analyses, we demonstrated that BMP9 regulates a distinct set of downstream target genes in MSCs [18,19,20,21,22,23,24], as well as cross-talking with other pathways [25,26,27,28,29]. Nonetheless, as one of the least studied BMPs, the essential mediators of BMP9-induced osteogenic differentiation in MSCs remain to be fully elucidated.
Notch signaling is known to function as an important regulator of bone formation [30]. Mammal Notch signaling consists of four Notch receptors (Notch1–4) and five Notch ligands (Dll1, Dll3, Dll4, Jagged1 and Jagged2) [30]. Notch signaling is activated by ligand binding to Notch receptor, followed by sequential proteolytic cleavages of Notch extracellular/transmembrane domains and releasing the Notch intracellular domain (NICD) [30, 31]. NICD is translocated into nucleus and interacts with DNA-binding protein CSL (CBF1/Suppressor of Hairless/LAG-1) and regulates downstream genes [30]. We previously showed that BMP9 synergizes with Notch signaling in osteogenic differentiation [32,33,34], although the exact role of Notch signaling in BMP9-induced osteogenesis remains to be fully understood.
Here, we investigate whether Notch signaling is necessary and/or sufficient to mediate BMP9-induced osteogenic differentiation in MSCs. By overexpressing dominant-negative Notch1 mutant or Notch ligands, and using Notch pathway genetically inactivated MSCs, we demonstrate that BMP9 activates Notch signaling at intermediate stage of osteogenic differentiation and that Notch signaling is required for effective bone formation induced by BMP9 in MSCs. Thus, our results strongly suggest that Notch signaling may play an essential role in coordinating BMP9-induced osteogenic differentiation of MSCs.
Materials and methods
Cell culture and chemicals
HEK-293 and C3H10T1/2 cells were from ATCC (Manassas, VA). The immortalized mouse embryonic fibroblasts (iMEFs) and immortalized mouse adipocyte-derived mesenchymal stem cells (iMADs) were previously described [35,36,37]. The cell lines were maintained in the conditions as described [38,39,40]. Compound E was purchased from Cayman Chemical (Ann Arbor, MI). Unless indicated otherwise, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Construction and generation of recombinant adenoviral vectors Ad-BMP9, Adr-dnNotch1, AdR-Dll1, AdR-Jag1, Ad-RFP and Ad-GFP
Recombinant adenoviruses were generated using AdEasy technology as described [41, 42]. The construction of Ad-BMP9 was previously described [12, 13]. The generation of AdR-dnNotch1, which expresses the extracellular domain with the transmembrane region (aa #l-aa #1705) of mouse Notch1, was also previously reported [32, 33]. For making AdR-Dll1 and AdR-Jag1, the coding regions of mouse Dll1 and Jagged1 were PCR amplified and subcloned into an adenoviral shuttle vector, and used to generate recombinant adenoviral vectors, resulting in pAdR-Dll1 and pAdR-Jag1, respectively, which were subsequently used to generate recombinant adenoviruses in HEK-293 or our recently engineered 293pTP or RAPA cells [43, 44]. Ad-BMP9 co-expresses enhanced GFP (eGFP), while AdR-dll1 and AdR-Jag1 co-express monomeric RFP (mRFP). Ad-GFP and Ad-RFP were used as mock virus controls as reported [29, 45, 46]. For all adenoviral infections, polybrene (4–8 µg/ml) was added to potentiate infection efficiency as previously described [47]. Detailed information about vector construction is available upon request.
Establishment of the stable expression of PS1 (DKO-PS1) in the presenilin 1 and 2 (PS1 and PS2) double-knockout mouse embryonic fibroblasts (MEFs) PS1 −/− /PS2 −/− (DKO)
MEFs derived from the PS1−/−/PS2−/− double-knockout mice (DKO) were previously reported [48]. The PS1−/−/PS2−/− DKO MEFs were infected with retroviral vector expressing PS1 or empty vector, and were selected in the presence of 4 μg/ml puromycin [48]. The resulting stable lines were designated as DKO-PS1 and DKO-EV, respectively. The expression of exogenous PS1 in DKO-PS1 cells was verified by Western blotting analysis.
RNA isolation, semi-quantitative RT-PCR (sqPCR) and touchdown quantitative PCR (TqPCR)
Total RNA was isolated using TRIZOL Reagents (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and subjected to reverse transcription reactions using hexamer and M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA). The cDNA products were further diluted 5- to 10-fold and used as PCR templates. The qPCR primers (Suppl. Table 1) were designed by using the Primer3 program. The sqPCR analysis was carried out as described [49,50,51,52,53]. Briefly, a touchdown cycling program was as follows: 94 °C for 2 min for 1 cycle; 92 °C for 20 s, 68 °C for 30 s, and 72 °C for 12 cycles decreasing 1 °C per cycle; and then at 92 °C for 20 s, 57 °C for 30 s, and 72 °C for 20 s for 20–25 cycles, depending on the abundance of a given gene. The PCR products were resolved on 1.5% agarose gels. Gapdh was used as the reference gene.
TqPCR analysis was carried out as previously described [54,55,56]. Briefly, the SYBR Green qPCR reactions (Bio-Rad Laboratories) were set up according to manufacturer’s instructions. TqPCR reactions were carried out in triplicate using the following conditions: 95 °C × 3′ for one cycle; 95 °C × 20′′, 66 °C × 10′′ for 4 cycles by decreasing 3 °C per cycle; then 95 °C × 20′′, 55 °C × 10′′, 70 °C × 1′′, followed by plate read for 40 cycles. Gapdh was used as a reference gene.
Alkaline phosphatase (ALP) activity assays
ALP activity was assessed quantitatively by a modified Great Escape SEAP Chemiluminescence assay (BD Clontech, Mountain View, CA) and/or qualitatively by histochemical staining assay (using a mixture of 0.1 mg/ml napthol AS-MX phosphate and 0.6 mg/ml Fast Blue BB salt) as described [26, 57]. For the chemilluminescence assays, each assay condition was performed in triplicate. The results were repeated in at least three independent experiments. The results were repeated in at least three independent batches of experiments. ALP activities were normalized by total cellular protein concentrations among the samples.
Alizarin Red S staining
Cells were seeded in 24-well cell culture plates and infected with the indicated adenoviruses. The cells were cultured in the presence of ascorbic acid (50 μg/mL) and β-glycerophosphate (10 mM) for 10–14 days. Mineralized matrix nodules were stained for calcium precipitation by means of Alizarin Red S staining as described previously [12, 27]. The staining of calcium mineral deposits was recorded under bright field microscopy.
Western blotting analysis
Total protein lysates from cultured cells were prepared essentially as previously described [48]. Cleared total cell lysate was denatured by boiling and resolved by 10% SDS–PAGE. After electrophoretic separation, proteins were transferred to Immobilon-P membranes, which were blocked and incubated overnight with primary antibodies against N-terminal of PS1 (or PS1NT, homemade) [58], nicastrin (goat, N-19, Santa Cruz Biotechnology), and β-actin (mAb, Cat# A5441, SigmaMillipore) as described [48]. After being washed, the membranes were incubated with a secondary antibody conjugated with horseradish peroxidase. Immune-reactive signals were detected using ECL kit (SigmaMillipore, America).
Immunohistochemical staining
The cells were fixed with 10% formalin, washed with PBS, and permeabilized with 1% NP-40. The fixed cells were blocked and incubated with an anti-osteocalcin (Ocn), or osteopontin (Opn) antibody (Santa Cruz Biotechnology). After being washed, cells were incubated with biotin-labeled secondary antibody for 30 min, followed by incubating cells with streptavidin-HRP conjugate for 20 min at room temperature. The presence of the expected protein was visualized by DAB staining and examined under a microscope. Stains with without primary antibody or control IgG were used as negative controls.
Immunofluorescence staining
Subconfluent C3H10T1/2 cells were infected with AdBMP9 or AdGFP. At 72 h post infection, the cells were fixed with 4% paraformaldehyde. The fixed cells were then treated with 0.1% Triton-100 and blocked with 10% bovine serum albumin. The cells were incubated with Notch1 antibody (against the NICD domain) (Santa Cruz Biotechnology) in 4 °C overnight and stained with Cy3-anti-mouse IgG secondary antibody (Jackson ImmunoResearch). Cell nuclei were counterstained with DAPI, followed by fluorescence microscopic imaging. Isotype IgG was used as a negative control. Staining reactions were done in triplicate.
Cell cycle analysis
Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and AdR-dnNotch1, AdR-Dll1 or AdR-Jag1. At 72 h post infection, the cells were collected, fixed, and stained with the Magic Solution for 30 min. The Hoechst 33258 stained cells were subjected to flow cytometric analysis using the BD FACSCalibur-HTS as described [59]. The acquired flow cytometry data were analyzed with the FlowJo v10.0 software. Each assay condition was done in triplicate.
Stem cell implantation and μCT analysis
Subconfluent DKO-PS1, DKO-EV, or iMEF cells were infected with Ad-BMP9 or Ad-GFP; or subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdRFP and AdR-dnNotch1, AdR-Dll1 or AdR-Jag1. At 24 h post infection, the cells were collected and resuspended in PBS for subcutaneous injection (5 × 106/injection) into the flanks of athymic nude (nu/nu) mice (five animals/group, 5–6-week-old, female, Harlan Sprague-Dawley). At 4wk post implantation, animals were sacrificed. Implantation sites were retrieved for μCT analysis, histologic evaluation, and other stains. All specimens were imaged using the μCT component of the GE Triumph (GE Healthcare, Piscataway, NJ, USA) trimodality preclinical imaging system. All image data analysis was performed using Amira 5.3 (Visage Imaging, Inc., San Diego, CA, USA); and 3D volumetric data and bone mean density hit maps were obtained as previously described [39, 60, 61].
Hematoxylin & eosin and trichrome staining
Retrieved tissues were fixed, decalcified in 10% formalin and embedded in paraffin. Serial sections of the embedded specimens were stained with hematoxylin and eosin (H & E). Masson Trichome staining was carried out as previously described [29, 62].
Statistical analysis
All quantitative experiments were performed in triplicate and/or repeated three independent batches of experiments. Data were expressed as mean ± SD. Statistical significances between treatments vs. control groups were determined by one-way analysis of variance and the Student’s t-test. A value of p < 0.05 was considered statistically significant.
Results
Blockade of Notch signaling diminishes BMP9-induced osteogenic differentiation of MSCs
We analyzed the endogenous expression profile of Notch receptors and ligands in MSCs. Semi-quantitative PCR revealed that the expression of three of the four Notch receptors and three of five Notch ligands were readily detected in C3H10T1/2 cells (Fig. 1a, panel a). Similar expression patterns of Notch receptors and ligands were obtained in iMEFs via qPCR analysis (Fig. 1a, panel b). When MSCs were stimulated with BMP9, we found the osteogenic marker ALP activity was inhibited by a Notch signaling inhibitor, the γ-secretase inhibitor Compound E (CE), in a dose-dependent fashion as assessed by ALP histochemical staining (Fig. 1b, panel a) and quantitative ALP measurement (Fig. 1b, panel b). Furthermore, the γ-secretase inhibitor CE was shown to effectively block BMP9-induced matrix mineralization in a dose-dependent fashion (Fig. 1c). These results suggest that inhibition of Notch signaling may significantly impair BMP9-induced osteogenic signaling.
BMP9-induced osteogenic differentiation of MSCs is inhibited by the γ-secretase inhibitor Compound E. (a) Most components of Notch signaling pathway express at a low level in MSCs. Total RNA was isolated from subconfluent C3H10T1/2 (a) and iMEFs (b) cells and subjected to semi-quantitative PCR (a) or TqPCR (b) analysis of the expression of Notch receptors and ligands. (b) The γ-secretase inhibitor Compound E (CE) inhibits BMP9-induced osteogenic marker ALP activity in MSCs. Subconfluent C3H10T1/2 cells were infected with AdBMP9 or AdGFP and treated with the indicated concentrations of CE. At 5 days of treatment, the cells were fixed and subjected to ALP histochemical staining (a). Alternatively, the cells were lysed at the indicated time points and subjected to chemiluminescence–based quantitative assay of ALP activity (b). Each assay condition was done in triplicate. *p < 0.05, **p < 0.001. (c) BMP9-induced matrix mineralization is effectively blunted by CE. Subconfluent C3H10T1/2 cells were infected with AdBMP9 or AdGFP, treated with the indicated concentrations of CE, and maintained in the mineralization medium for 14 days. The cells were fixed and subjected to Alizarin Red S staining. Each assay condition was done in triplicate. Representative results are shown
In order to specifically block Notch signaling, we constructed and generated an adenoviral vector that expresses the dominant-negative extracellular domain of Notch1 (dnNotch1) (Fig. 2a). We showed the adenoviral vector AdRdnNotch1 infected cells effectively and expressed a high level of dnNotch1 (Fig. 2b). The dominant-negative Notch1 was shown to effectively inhibit BMP9-induced ALP activity in C3H10T1/2 cells (Fig. 2c), and such inhibitory effect was shown to be dose-dependent (Fig. 2d). Furthermore, BMP9-induced matrix mineralization was inhibited by dnNotch1 in MSCs (Fig. 2e). Thus, these results indicate that a blockade of Notch signaling would significantly impair BMP9-initiated osteogenic signaling in MSCs.
Dominant-negative mutant Notch1 inhibits BMP9-induced osteogenic differentiation of MSCs. a Schematic representation of the dominant-negative Notch1 (dnNotch1), which contains the extracellular domain and the transmembrane region of mouse Notch1. b Adenovirus-mediated overexpression of dnNocth1. Subconfluent C3H10T1/2 cells were infected with AdR-dnNotch1 or AdRFP control virus for 36 h. Total RNA was collected and subjected to semi-quantitative PCR (sqPCR) using primers for mouse Notch1 extracellular domain or mouse Gapdh. The sqPCR was done in triplicate; and representative results are shown. c, d dnNotch1 inhibits BMP9-induced ALP activity in MSCs. Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and different titers of AdR-dnNotch1. At five days after infection, the ALP activity of the infected cells were determined either by qualitative histochemical staining (a) or quantitative chemiluminescence assay (b). Each assay condition was done in triplicate. Representative results are shown. *p < 0.05, **p < 0.001. e dnNotch1 diminishes BMP9-induced matrix mineralization. Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and/or AdRdnNotch1 and maintained in the mineralization medium for 14 days. The cells were fixed and subjected to Alizarin Red S staining. Each assay condition was done in triplicate. Representative results are shown
Genetic inactivation of Notch signaling pathway significantly diminishes BMP9-induced osteogenic differentiation of MSCs
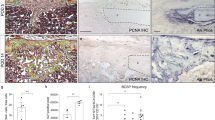
To further demonstrate the essential role of Notch signaling in BMP9-induced osteogenic differentiation, we established the Presenilin 1 and Presenilin 2 PS1−/−/PS2−/− DKO MEFs that were stably transduced with retroviral vector expressing Flag-tagged PS1 (DKO-PS1) or empty vector (DKO-EV) (Fig. 3a). When these cells were stimulated with BMP9, we found that the ALP activity was readily detected in the PS1-reconstituted DKO-PS1 cells while only minimal ALP activity was detected in the DKO-EV cells (Fig. 3b). Quantitative analysis of BMP9-induced ALP activities in the DKO-PS1 and DKO-EV cells obtained similar results at multiple time points (Fig. 3c, panel a). Furthermore, the diminished ALP activities in DKO-EV cells were not reversible even at high titers of AdBMP9 infection while there was an apparent dose-dependent response in DKO-PS1 cells (Fig. 3c, panel b).
Genetic inactivation of Notch signaling pathway significantly diminishes BMP9-induced osteogenic differentiation of MSCs. (a) Restoration of PS1 expression in PS1−/−/PS2−/− MEFs. The PS1−/−/PS2−/− double-knockout (DKO) MEFs were stably transduced with retroviral vector expressing Flag-tagged PS1 (DKO-PS1) or empty vector (DKO-EV). Sunconfluent stable line cells were lysed and subjected to Western blotting analysis using antibodies against the N-terminal fragment of PS1 (PS1-NTF) (a), nicastrin (b) or β-actin (c). Each assay was done in two independent batches of experiments. Representative results are shown. b, c Double-knockout PS1−/−/PS2−/− significantly blunts BMP9-induced ALP activity in MSCs. Subconfluent DKO-EV or DKO-PS1 cells were infected with AdBMP9 or AdGFP. ALP activity of the infected cells were determined either by qualitative histochemical staining at day 5 (b) or by quantitative chemiluminescence assay at 3, 5, or 7 days after infection (c, panel a). ALP activity for different titers of AdBMP9 infected DKO-EV or DKO-PS1 cells, was also determined at three days after infection (c, panel b). Each assay condition was done in triplicate. Representative results are shown. **p < 0.001. d Double-knockout PS1−/−/PS2−/− significantly inhibits BMP9-induced expression of late osteogenic markers in MSCs. Subconfluent DKO-EV or DKO-PS1 cells were infected with AdBMP9 or AdGFP for five days. Cells were fixed and subjected to immunocytochemical staining with the osteocalcin (Ocn) (a) or osteopontin (Opn) (b) antibody. Minus primary antibody staining was used as a negative control (data not shown). Each staining condition was done in duplicate. Representative results are shown. e Double-knockout PS1−/−/PS2−/− significantly inhibits BMP9-induced matrix mineralization in MSCs. Subconfluent DKO-EV or DKO-PS1 cells were infected with AdBMP9 or AdGFP and maintained in the mineralization medium for 14 days. The cells were fixed and subjected to Alizarin Red S staining. Each assay condition was done in triplicate. Representative results are shown
We further analyzed the BMP9-induced expression of late osteogenic markers osteocalcin (Ocn) and osteopontin (Opn). We found that BMP9-induced Ocn expression was diminished in DKO-EV cells, compared with that in DKO-PS1 cells (Fig. 3d, panel a). Similarly, BMP9-induced Opn expression was almost undetectable in DKO-EV cells, but readily detected in DKO-PS1 cells (Fig. 3d, panel b). Lastly, we found that BMP9-induced matrix mineralization was significantly diminished in DKO-EV cells, compared that in DKO-PS1 cells (Fig. 3e). Collectively, the above results further demonstrate that Notch signaling plays an essential role in BMP9-induced osteogenic differentiation in MSCs.
Exogenous expression of Notch ligands promotes BMP9-induced osteogenic differentiation of MSCs
We also examined if the activation of Notch signaling via overexpressing Notch ligands affected BMP9-induced osteogenic activity, and constructed adenoviral vectors overexpressing Dll1 and Jag1, or AdR-Dll1 and AdR-Jag1, respectively. We showed that these adenoviral vectors mediated high levels of expression of both Dll1 and Jag1 in MSCs (Fig. 4a, panel a and b). When MSCs were co-infected with AdBMP9 and AdR-Dll1, the BMP9-induced ALP activities were significantly enhanced while overexpression of Dll1 alone stimulated negligible ALP activity (Fig. 4b, panel a). Similarly, the exogenous expression of Jag1 effectively enhanced BMP9-induced ALP activities (Fig. 4b, panel b). Furthermore, overexpression of either Dll1 or Jag-1 was shown to significantly enhance BMP9-induced matrix mineralization in MSCs (Fig. 4c, panel a and b). Therefore, the above results strongly suggest that the activation of Notch signaling may facilitate BMP9-induced osteogenic differentiation of MSCs.
Exogenous expression of Notch ligands Dll11 and Jag1 enhances BMP9-induced osteogenic differentiation of MSCs. a Adenovirus-mediated overexpression of Notch ligands Dll1 and Jag1. Subconfluent C3H10T1/2 cells were infected with AdR-Dll1, AdR-Jag1, or AdRFP control virus for 36 h. Total RNA was collected and subjected to sqPCR analysis using primers for mouse Dll1, Jag1, or Gapdh. The sqPCR was done in triplicate and representative results are shown. b Overexpression of Dll1 or Jag1 enhances BMP9-induced ALP activity in MSCs. Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and AdR-Dll1 (a) or AdR-Jag1 (b). At 3, 5, and 7 days after infection, the ALP activity of the infected cells were determined by quantitative chemiluminescence assay. Each assay condition was done in triplicate. Representative results are shown. *p < 0.05, **p < 0.001. c Overexpression of Dll1 or Jag1 augments BMP9-induced matrix mineralization. Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and/or AdR-Dll1 (a) or AdR-Jag1 (b) and maintained in the mineralization medium for 14 days. The cells were fixed and subjected to Alizarin Red S staining. Each assay condition was done in triplicate. Representative results are shown
Blockade of Notch signaling significantly diminishes BMP9-induced ectopic bone formation
We tested the in vivo effect of Notch signaling inhibition on BMP9-induced bone formation from MSCs. When iMEF cells were co-infected with AdBMP9 and AdR-dnNotch1, AdR-Dll1, or AdR-Jag1, we found that the expression of dnNotch1 significantly inhibited the volumetric values of BMP9-induced bone formation (p < 0.001) (Fig. 5a, panel a vs. b and Fig. 5b, panel a), while overexpression of Dll1 or Jag1 slightly increased the average bone volume although there was no statistical significance (Fig. 5a, panel a vs. c and d and Fig. 5b, panel, a). Furthermore, overexpression of dnNotch1 significantly inhibited the mean bone density of the formed ectopic bone (p < 0.001) while the exogenous expression of Dll1 and Jag1 did not significantly affect the mean bone density (Fig. 5b, panel b). Histological evaluation and trichrome staining of the retrieved masses indicated that dnNotch1 overexpression significantly inhibited BMP9-induced bone formation as evidenced by significantly decreased trabecular bone and reduced mineralized bone matrix, compared with that of RFP control group (Fig. 5c, panel a and b). Interestingly, overexpression of Dll1 or Jag1 only slightly increased BMP9-induced trabecular bone formation and bone matrix mineralization (Fig. 5c, panel a and b). Nonetheless, these results are consistent with the in vitro findings, in which a blockade of Notch signaling leads to inhibition of BMP9-induced osteogenic differentiation.
The inhibition of Notch signaling by dnNotch1 diminishes BMP9-induced ectopic bone formation, while exogenous expression of Notch ligands enhances BMP9-induced ectopic bone formation in vivo. a, b Subconfluent iMEF cells were co-infected with AdBMP9 or AdRFP and AdR-dnNotch1, AdR-Dll1, or AdR-Jag1 for 24 h, and collected for subcutaneous injection into the flanks of athymic nude mice. At 4 weeks after implantation, bony masses were retrieved from BMP9 treatment groups while no masses were recovered from the non-BMP9 treatment groups. The retrieved masses were fixed and subjected to µCT imaging (a, panels a–d), and the imaging data were further analyzed for average bone volume (b, panel a) and mean bone density (b, panel b). **p < 0.001. c Histological and special staining. The retrieved masses were fixed, decalcified and subjected to H & E staining (a) and Trichrome staining (b). Representative results are shown
We further examined the bone forming capability in DKO-EV and DKO-PS1 cells upon BMP9 stimulation. While these cells were equally transduced with AdBMP9 (Fig. 6a, panels a and b), the DKO-EV cells formed dramatically much smaller bony masses, compared with that in the DKO-PS1 cells (p < 0.001) (Fig. 6a, panels c, d, e). Furthermore, the average bone density was significantly decreased in DKO-EV cells, compared with that in DKO-PS1 cells (p < 0.001) (Fig. 6a, panel f). Histological analysis confirms that BMP9 induced significantly smaller bone mass with numerous undifferentiated cells in the center of the mass in DKO-EV cells, compared with that in DKO-PS1 cells (Fig. 6b, panels a and b vs. c and d). Trichrome staining indicates BMP9 induced much fewer mature trabecular bone in the DKO-EV cells than that in DKO-PS1 cells (Fig. 6c, panels a and b vs. c and d). Collectively, these in vivo results further demonstrate that Notch signaling plays an essential role in BMP9-induced osteogenic differentiation and bone formation.
Genetic inactivation of Notch signaling blunts BMP9-induced ectopic bone formation in vivo. a Double-knockout PS1−/−/PS2−/− significantly blunts BMP9-induced ectopic bone formation in MSCs. Subconfluent DKO-EV or DKO-PS1 cells were infected with AdBMP9 or AdGFP for 24 h (a, b) and collected for subcutaneous injection into the flanks of athymic nude mice. At 4 weeks after implantation, bony masses were retrieved from BMP9 treatment groups while no masses were recovered from the GFP treatment group (c). The retrieved masses were fixed and subjected to µCT imaging (d), and the imaging data were further analyzed for average bone volume (e) and mean bone density (f). **p < 0.001. b H & E staining. The retrieved masses from BMP9-treated DKO-EV cells (a, b) and BMP9-treated DKO-PS1 cells (c, d) were decalcified and subjected to H & E staining. Representative results are shown. c Trichrome staining. The retrieved masses from BMP9-treated DKO-EV cells (a, b) and BMP9-treated DKO-PS1 cells (c, d) were decalcified and subjected to Trichrome staining. Representative results are shown
BMP9 upregulates Notch signaling at the intermediate stage of osteogenic differentiation of MSCs, while notch signaling enhances BMP9-stimulated progenitor cell proliferation
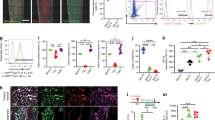
To understand if BMP9 regulates Notch signaling in MSCs, we analyzed the expression status of BMP9-induced early-responsive genes in DKO-EV and DKO-PS1 cells. We found that Smad6, Smad7, and Id2 were significantly upregulated by BMP9 at day 2 in both DKO-EV and DKO-PS1 cells (Fig. 7a), indicating that BMP9-induced the expression of these early-responsive genes is independent of and possibly upstream of Notch signaling.
BMP9 upregulates Notch signaling at the intermediate stage of osteogenic differentiation of MSCs, while Notch signaling enhances BMP9-stimulated progenitor cell proliferation. a Inactivation of Notch signaling does not significantly impair BMP9-induced expression of immediate early genes. Subconfluent DKO-EV or DKO-PS1 cells were infected with AdBMP9 or AdGFP. Total RNA was collected at the indicated time points and subjected to TqPCR with primers specific for mouse Smad6, Smad7, and Id2. Mouse Gapdh was used as a reference gene. The TqPCR assays were done in triplicate. The fold of relative expression was calculated by dividing the normalized expression in BMP9-treated group by that in the GFP group. *p < 0.05, **p < 0.001. b BMP9 upregulates Notch signaling at the intermediate stage of osteogenic differentiation of MSCs. Subconfluent iMAD cells were infected with AdBMP9 or AdGFP. Total RNA was collected at the indicated time points and subjected to TqPCR with primers specific for mouse Notch receptors (a) and ligands (b). Mouse Gapdh was used as a reference gene. The TqPCR assays were done in triplicate. The fold of relative expression was calculated by dividing the normalized expression in BMP9-treated group by that in the GFP group. *p < 0.05, **p < 0.001. c BMP9 induces nuclear accumulation of NICD in MSCs. Subconfluent C3H10T1/2 cells were infected with AdBMP9 or AdGFP. At 72 h post infection, the cells were fixed and subjected to immunofluorescence staining with an antibody against the Notch1 intracellular domain (Santa Cruz Biotechnology). Isotype IgG was used as a negative control. Cell nuclei were counterstained with DAPI. Staining reactions were done in triplicate. Representative results are shown. d Notch signaling promotes BMP9-regulated cell cycle progression in MSCs. Subconfluent C3H10T1/2 cells were co-infected with AdBMP9 or AdGFP and AdR-dnNotch1, AdR-Dll1 or AdR-Jag1. At 72 h post infection, the cells were fixed, stained with Hoechst 33258 and subjected to flow cytometric analysis. Percentages of the cells in the S/G2 phases were calculated using the FlowJo software. Each assay wa performed in triplicate
We further analyzed the expression profiles of Notch receptors and ligands in iMAD cells upon BMP9 stimulation and found that Notch receptors, most notably Notch1 and Notch4, were significantly upregulated at 5 days after BMP9 stimulation while Notch3 was upregulated at 3 days after BMP9 transduction (Fig. 7b, panel a), corresponding to the intermediate stage of BMP9-induced osteogenic differentiation in vitro [5, 11]. Similarly, we found that BMP9 significantly upregulated the expression of Jag1, Jag2, Dll1, Dll2, and Dll3 at 5 days after BMP9 stimulation while Dll1 showed a small increase at day 3 (Fig. 7b, panel b). The upregulation timeline also corresponded to the intermediate stage of BMP9-induced osteogenic differentiation in vitro.
We also determined whether the downstream event of Notch signaling is activated by BMP9 by examining the production of Notch intracellular domain (NICD). When MSCs were infected with AdBMP9 for 72 h, NICD-specific immunostaining detected significantly elevated nuclear localization of NICD, compared with that of the control groups (Fig. 7c), indicating that activation of Notch signaling is implicated in BMP9-induced osteogenic signaling in MSCs. Furthermore, cell cycle analysis revealed that BMP9 treatment stimulated MSC proliferation and increased the percentage of cells in S/G2 phases, e.g., from 39.83% (GFP group) to 49.83% (Fig. 7d, panels a vs. b). Overexpression of dnNotch1 itself did not significantly inhibit cell proliferation (Fig. 7d, panels a vs. c), but was shown to inhibit BMP9-induced proliferation, e.g., 49.83% cells in S/G2 (BMP9 alone) vs. 44.17% in S/G2 phases (BMP9 + dnNotch1) (Fig. 7d, panels b vs. d). Conversely, overexpression of Notch ligands Dll1 and Jag1 significantly promoted BMP9-induced percentage of cells in S/G2 phases, e.g., from 49.83% (BMP9 alone) to 56.33% and 53.20%, respectively (Fig. 7d, panels b vs. e and f). Collectively, these results strongly suggest that Notch signaling may participate in BMP9-induced the proliferative expansion of progenitors and the eventual osteogenic terminal differentiation of MSCs. Thus, Notch signaling may play an essential role in coordinating BMP9-induced potent osteogenic differentiation of MSCs.
Discussion
We previously identified BMP9 as one of the most potent osteogenic BMPs and later found that BMP9 also promotes effective adipogenic differentiation of MSCs [11,12,13, 63]. Mechanistically, we demonstrated that TGFβ type I receptors ALK1 and ALK2 are required for BMP9 binding to trigger osteogenic signaling [57], which involves in regulating cascade events leading to bone formation MSCs [18,19,20,21,22,23,24,25,26,27,28,29]. Nonetheless, as BMP9 is one of the least studied BMPs, the exact molecular mechanism underlying BMP9-induced osteogenesis remains to be defined.
On the other hand, Notch signaling is known to be involved in bone and skeletal development [64]. For example, Jagged1 expression was able to induce osteogenesis [65], whereas the genetic deletion of Jagged1 resulted in defects in osteoblast development and differentiation during maxillary ossification [66]. We found that a Notch downstream target gene Hey1 was upregulated by BMP9 [21], suggesting BMP9 may act upstream of Notch signaling although, unlike BMP9, an activation of Notch signaling alone (e.g., by overexpressing either NICD or Notch ligands) fails to induce de novo osteogenic differentiation and ectopic bone formation from MSCs [33]. Nonetheless, we have recently demonstrated that Notch signaling can synergize with BMP9 in inducing bone formation [33]. Our current studies strongly suggest that Notch signaling is a critical mediator of BMP9-induced osteogenic differentiation.
It remains to be fully elucidated about how BMP9 regulates Notch signaling in the context of bone formation. Our results indicate that Notch receptors and/or ligands are not upregulated at the immediate early stage of BMP9 stimulation, suggesting that BMP9 may regulate Notch receptors and/or ligands in an indirect fashion. Interestingly, we have recently shown that lncRNA H19 may play an important role in mediating BMP9-induced osteogenic signaling in MSCs [34]. Although H19 was identified nearly three decades ago [67], the diverse biological functions of H19 remain to be fully understood [68].
One aspect of H19 actions may regulate microRNA (miRNA) functions by serving as a molecular sponge. In fact, it has been recently demonstrated that H19 modulates let-7 availability by acting as a molecular sponge [69]. We found H19 is significantly upregulated in MSCs upon BMP9 stimulation [34]. We investigated whether H19 modulates the expression levels of a panel of miRNAs that are predicted to modulate Notch ligands and receptors upon BMP9 stimulation, and found that Notch-associated miRNAs (e.g., miR-107, miR-27b, miR-106b, miR125a, and miR17) may be modulated by H19 in response to BMP9 stimulation in MSCs. Thus, it is conceivable that BMP9 may regulate Notch signaling through directly modulating lncRNAs and/or miRNAs, which target the essential components of Notch signaling pathway, leading to coordinated regulation of osteogenic differentiation. Nonetheless, the detailed mechanisms remain to be fully understood.
References
Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74.
Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7.
Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220.
Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–94.
Luu HH, Song WX, Luo X, et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 2007;25:665–77.
Deng ZL, Sharff KA, Tang N, et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 2008;13:2001–21.
Zhang J, Li L. BMP signaling and stem cell regulation. Dev Biol. 2005;284:1–11.
Wang RN, Green J, Wang Z, et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105.
Zhang F, Song J, Zhang H, et al. Wnt and BMP Signaling Crosstalk in Regulating Dental Stem Cells: Implications in Dental Tissue Engineering. Genes Dis. 2016;3:263–76.
Luther G, Wagner ER, Zhu G, et al. BMP-9 induced osteogenic differentiation of mesenchymal stem cells: molecular mechanism and therapeutic potential. Curr Gene Ther. 2011;11:229–40.
Lamplot JD, Qin J, Nan G, et al. BMP9 signaling in stem cell differentiation and osteogenesis. Am J Stem Cells. 2013;2:1–21.
Cheng H, Jiang W, Phillips FM, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J Bone Jt Surg Am. 2003;85-A:1544–52.
Kang Q, Sun MH, Cheng H, et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–20.
Song JJ, Celeste AJ, Kong FM, et al. Bone morphogenetic protein-9 binds to liver cells and stimulates proliferation. Endocrinology. 1995;136:4293–7.
Lopez-Coviella I, Berse B, Krauss R, et al. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science. 2000;289:313–6.
Chen C, Grzegorzewski KJ, Barash S, et al. An integrated functional genomics screening program reveals a role for BMP-9 in glucose homeostasis. Nat Biotechnol. 2003;21:294–301.
Li W, Salmon RM, Jiang H, et al. Regulation of the ALK1 ligands, BMP9 and BMP10. Biochem Soc Trans. 2016;44:1135–41.
Peng Y, Kang Q, Cheng H, et al. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J Cell Biochem. 2003;90:1149–65.
Peng Y, Kang Q, Luo Q, et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–9.
Luo Q, Kang Q, Si W, et al. Connective Tissue Growth Factor (CTGF) Is Regulated by Wnt and Bone Morphogenetic Proteins Signaling in Osteoblast Differentiation of Mesenchymal Stem Cells. J Biol Chem. 2004;279:55958–68.
Sharff KA, Song WX, Luo X, et al. Hey1 Basic helix-loop-helix protein plays an important role in mediating BMP9-induced Osteogenic Differentiation of Mesenchymal Progenitor Cells. J Biol Chem. 2009;284:649–59.
Huang E, Zhu G, Jiang W, et al. Growth hormone synergizes with BMP9 in osteogenic differentiation by activating the JAK/STAT/IGF1 pathway in murine multilineage cells. J Bone Miner Res. 2012;27:1566–75.
Wang Y, Hong S, Li M, et al. Noggin resistance contributes to the potent osteogenic capability of BMP9 in mesenchymal stem cells. J Orthop Res. 2013;31:1796–803.
Hu N, Jiang D, Huang E, et al. BMP9-regulated angiogenic signaling plays an important role in the osteogenic differentiation of mesenchymal progenitor cells. J Cell Sci. 2013;126:532–41.
Tang N, Song WX, Luo J, et al. BMP9-induced osteogenic differentiation of mesenchymal progenitors requires functional canonical Wnt/beta-catenin signaling. J Cell Mol Med. 2009;13:2448–64.
Chen L, Jiang W, Huang J, et al. Insulin-like growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic differentiation and bone formation. J Bone Miner Res. 2010;25:2447–59.
Zhang W, Deng ZL, Chen L, et al. Retinoic acids potentiate BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2010;5:e11917.
Liu X, Qin J, Luo Q, et al. Cross-talk between EGF and BMP9 signalling pathways regulates the osteogenic differentiation of mesenchymal stem cells. J Cell Mol Med. 2013;17:1160–72.
Zhang H, Wang J, Deng F, et al. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs). Biomaterials. 2015;39:145–54.
Zanotti S, Canalis E. Notch signaling and the skeleton. Endocr Rev. 2016;37:223–53.
Yavropoulou MP, Yovos JG. The role of Notch signaling in bone development and disease. Horm (Athens). 2014;13:24–37.
Li R, Zhang W, Cui J, et al. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr Cancer Drug Targets. 2014;14:274–85.
Liao J, Wei Q, Zou Y, et al. Notch signaling augments BMP9-induced bone formation by promoting the osteogenesis-angiogenesis coupling process in mesenchymal stem cells (MSCs). Cell Physiol Biochem. 2017;41:1905–23.
Liao J, Yu X, Hu X, et al. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget. 2017;8:53581–601.
Huang E, Bi Y, Jiang W, et al. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PLoS ONE. 2012;7:e32428.
Wang N, Zhang W, Cui J, et al. The piggyBac transposon-mediated expression of SV40 T antigen efficiently immortalizes mouse embryonic fibroblasts (MEFs). PLoS One. 2014;9:e97316.
Lu S, Wang J, Ye J, et al. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am J Transl Res. 2016;8:3710–30.
Fan J, Wei Q, Liao J, et al. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–19.
Wang J, Liao J, Zhang F, et al. NEL-Like Molecule-1 (Nell1) is regulated by bone morphogenetic protein 9 (BMP9) and potentiates BMP9-induced osteogenic differentiation at the expense of adipogenesis in mesenchymal stem cells. Cell Physiol Biochem. 2017;41:484–500.
Zhang F, Li Y, Zhang H, et al. Anthelmintic mebendazole enhances cisplatin’s effect on suppressing cell proliferation and promotes differentiation of head and neck squamous cell carcinoma (HNSCC). Oncotarget. 2017;8:12968–82.
Luo J, Deng ZL, Luo X, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–47.
Lee CS, Bishop ES, Zhang R, et al. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63.
Wu N, Zhang H, Deng F, et al. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–37.
Wei Q, Fan J, Liao J, et al. Engineering the rapid adenovirus production and amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell Physiol Biochem. 2017;41:2383–98.
Ye J, Wang J, Zhu Y, et al. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed Mater. 2016;11:025021.
Yan Z, Yin L, Wang Z, et al. A novel organ culture model of mouse intervertebral disc tissues. Cells Tissues Organs. 2016;201:38–50.
Zhao C, Wu N, Deng F, et al. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS ONE. 2014;9:e92908.
Vetrivel KS, Zhang X, Meckler X, et al. Evidence that CD147 modulation of beta-amyloid (Abeta) levels is mediated by extracellular degradation of secreted Abeta. J Biol Chem. 2008;283:19489–98.
Huang J, Bi Y, Zhu GH, et al. Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells. Liver Int. 2009;29:1569–81.
Zhu GH, Huang J, Bi Y, et al. Activation of RXR and RAR signaling promotes myogenic differentiation of myoblastic C2C12 cells. Differentiation. 2009;78:195–204.
Bi Y, He Y, Huang J, et al. Functional characteristics of reversibly immortalized hepatic progenitor cells derived from mouse embryonic liver. Cell Physiol Biochem. 2014;34:1318–38.
Bi Y, Huang J, He Y, et al. Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J Cell Biochem. 2009;108:295–303.
Chen X, Cui J, Yan Z, et al. Sustained high level transgene expression in mammalian cells mediated by the optimized piggyBac transposon system. Genes Dis. 2015;2:96–105.
Zhang Q, Wang J, Deng F, et al. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR Green qPCR. PLoS ONE. 2015;10:e0132666.
Liao J, Wei Q, Fan J, et al. Characterization of retroviral infectivity and superinfection resistance during retrovirus-mediated transduction of mammalian cells. Gene Ther. 2017;24:333–41.
Deng Y, Wang Z, Zhang F, et al. A Blockade of IGF signaling sensitizes human ovarian cancer cells to the anthelmintic niclosamide-induced anti-proliferative and anticancer activities. Cell Physiol Biochem. 2016;39:871–88.
Luo J, Tang M, Huang J, et al. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J Biol Chem. 2010;285:29588–98.
Thinakaran G, Regard JB, Bouton CM, et al. Stable association of presenilin derivatives and absence of presenilin interactions with APP. Neurobiol Dis. 1998;4:438–53.
Deng Y, Zhang J, Wang Z, et al. Antibiotic monensin synergizes with EGFR inhibitors and oxaliplatin to suppress the proliferation of human ovarian cancer cells. Sci Rep. 2015;5:17523.
Li Y, Wagner ER, Yan Z, et al. The calcium-binding protein S100A6 accelerates human osteosarcoma growth by promoting cell proliferation and inhibiting osteogenic differentiation. Cell Physiol Biochem. 2015;37:2375–92.
Dumanian ZP, Tollemar V, Ye J, et al. Repair of critical sized cranial defects with BMP9-transduced calvarial cells delivered in a thermoresponsive scaffold. PLoS One. 2017;12:e0172327.
Wang J, Zhang H, Zhang W, et al. Bone morphogenetic protein-9 (BMP9) effectively induces osteo/odontoblastic differentiation of the reversibly immortalized stem cells of dental apical papilla. Stem Cells Dev. 2014;23:1405–16.
Kang Q, Song WX, Luo Q, et al. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18:545–59.
Canalis E. Notch signaling in osteoblasts. Sci Signal. 2008;1:pe17.
Dishowitz MI, Zhu F, Sundararaghavan HG, et al. Jagged1 immobilization to an osteoconductive polymer activates the Notch signaling pathway and induces osteogenesis. J Biomed Mater Res A. 2014;102:1558–67.
Hill CR, Yuasa M, Schoenecker J, et al. Jagged1 is essential for osteoblast development during maxillary ossification. Bone. 2014;62:10–21.
Brannan CI, Dees EC, Ingram RS, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36.
Matouk IJ, Halle D, Raveh E, et al. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget. 2016;7:3748–65.
Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12.
Acknowledgements
The authors thank Drs. Wim Annaert and Bart De Strooper of Katholieke Universiteit Leuven and Flanders Interuniversity Institute for Biotechnology, Belgium, for kind provision of the PS1/2 DKO MEFs. The reported work was supported in part by research grants from the National Institutes of Health (CA226303, DE020140 to TCH and RRR), the US Department of Defense (OR130096 to JMW), the Scoliosis Research Society (TCH and MJL), the National Key Research and Development Program of China (2016YFC1000803 and 2011CB707906 to TCH), the National Natural Science Foundation of China (#81401536 to WWZ; and #81301551 to EH), the Foundation for Outstanding Young Scientist of Shandong Province (#BS2014YY035 to WWZ), and the Science and Technology Research Project of Chongqing Municipal Education Commission of China (KJ1500217 to JC). Wenwen Zhang, Enyi Huang, and Junyi Liao received scholarship funding from the China Scholarship Council. This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number UL1 TR000430. Funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cui, J., Zhang, W., Huang, E. et al. BMP9-induced osteoblastic differentiation requires functional Notch signaling in mesenchymal stem cells. Lab Invest 99, 58–71 (2019). https://doi.org/10.1038/s41374-018-0087-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0087-7
This article is cited by
-
Identification of genomic characteristics and selective signals in Guizhou black goat
BMC Genomics (2024)
-
Modeling lung diseases using reversibly immortalized mouse pulmonary alveolar type 2 cells (imPAC2)
Cell & Bioscience (2022)
-
TAZ promotes osteogenic differentiation of mesenchymal stem cells line C3H10T1/2, murine multi-lineage cells lines C2C12, and MEFs induced by BMP9
Cell Death Discovery (2022)
-
Regulation of osteogenic differentiation by the pro-inflammatory cytokines IL-1β and TNF-α: current conclusions and controversies
Human Cell (2022)
-
Hydrogel contained valproic acid accelerates bone-defect repair via activating Notch signaling pathway in ovariectomized rats
Journal of Materials Science: Materials in Medicine (2022)