Abstract
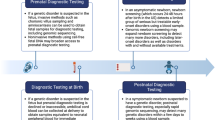
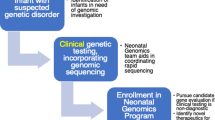
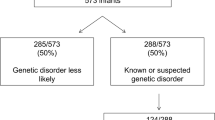
Genetic disorders presenting in the neonatal period can have a significant impact on morbidity and mortality. Early diagnosis can facilitate timely prognostic counseling to families and possibility of precision care, which could improve outcome. As availability of diagnostic testing expands, the required knowledge base of the neonatologist must also expand to include proper application and understanding of genetic testing modalities, especially where availability of clinical genetics consultation is limited. Herein, we review genetic tests utilized in the neonatal intensive care unit (NICU) providing background on the technology, clinical indications, advantages, and limitations of the tests. This review will span from classic cytogenetics to the evolving role of next generation sequencing and its impact on the management of neonatal disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Yang L, Liu X, Li Z, Zhang P, Wu B, Wang H, et al. Genetic aetiology of early infant deaths in a neonatal intensive care unit. J Med Genet. 2019. https://doi.org/10.1136/jmedgenet-2019-106221.
McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children’s hospital. Am J Hum Genet. 2004;74:121–7.
French CE, Delon I, Dolling H, Sanchis-Juan A, Shamardina O, Mégy K, et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45:627–36.
Petrikin JE, Willig LK, Smith LD, Kingsmore SF. Rapid whole genome sequencing and precision neonatology. Semin Perinatol. 2015;39:623–31.
Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re3.
Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 2018;3:10.
Tjio JH, Levan A. The chromosome number of man. Hereditas. 1956;42:1–6.
Pasquier L, Fradin M, Chérot E, Martin-Coignard D, Colin E, Journel H, et al. Karyotype is not dead (yet)! Eur J Med Genet. 2016;59:11–15.
Huber D, Voith von Voithenberg L, Kaigala GV. Fluorescence in situ hybridization (FISH): history, limitations and what to expect from micro-scale FISH? Micro Nano Eng. 2018;1:15–24.
Cui C, Shu W, Li P. Fluorescence in situ hybridization: cell-based genetic diagnostic and research applications. Front Cell Dev Biol. 2016;4. https://doi.org/10.3389/fcell.2016.00089.
McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JAS, et al. 22q11.2 deletion syndrome. Nat Rev Dis Prim. 2015;1:1–19.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2019. https://doi.org/10.1038/s41436-019-0686-8.
Manning M, Hudgins L, Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–5.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Wang J-C, Ross L, Mahon LW, Owen R, Hemmat M, Wang BT, et al. Regions of homozygosity identified by oligonucleotide SNP arrays: evaluating the incidence and clinical utility. Eur J Hum Genet. 2015;23:663–71.
Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–67.
Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, et al. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139:481–93.
Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–7.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51.
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6.
Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46.
Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem. 2013;6:287–303.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8.
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–7.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Lincoln SE, Truty R, Lin C-F, Zook JM, Paul J, Ramey VH, et al. A rigorous interlaboratory examination of the need to confirm next-generation sequencing–detected variants with an orthogonal method in clinical genetic testing. J Mol Diagn. 2019;21:318–29.
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47.
Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55.
Baruch S, Hudson K. Civilian and military genetics: nondiscrimination policy in a post-GINA world. Am J Hum Genet. 2008;83:435–44.
Hellwig LD, Turner C, Manolio TA, Haigney M, James CA, Murray B, et al. Return of secondary findings in genomic sequencing: Military implications. Mol Genet Genom Med. 2019;7:e00483.
De Castro M, Biesecker LG, Turner C, Brenner R, Witkop C, Mehlman M, et al. Genomic medicine in the military. NPJ Genom Med. 2016;1:15008.
ACMG Board of Directors Points to consider for informed consent for genome/exome sequencing. Genet Med. 2013;15:748–9.
Shellhaas RA, Wusthoff CJ, Tsuchida TN, Glass HC, Chu CJ, Massey SL, et al. Profile of neonatal epilepsies: characteristics of a prospective US cohort. Neurology. 2017;89:893–9.
Møller RS, Larsen LHG, Johannesen KM, Talvik I, Talvik T, Vaher U, et al. Gene panel testing in epileptic encephalopathies and familial epilepsies. Mol Syndromol. 2016;7:210–9.
Kapoor RR, Flanagan SE, Arya VB, Shield JP, Ellard S, Hussain K. Clinical and molecular characterisation of 300 patients with congenital hyperinsulinism. Eur J Endocrinol. 2013;168:557–64.
Stanley CA. Perspective on the genetics and diagnosis of congenital hyperinsulinism disorders. J Clin Endocrinol Metab. 2016;101:815–26.
Togawa T, Sugiura T, Ito K, Endo T, Aoyama K, Ohashi K, et al. Molecular genetic dissection and neonatal/infantile intrahepatic cholestasis using targeted next-generation sequencing. J Pediatr. 2016;171:171–7.e1–4.
Daoud H, Luco SM, Li R, Bareke E, Beaulieu C, Jarinova O, et al. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. Can Med Assoc J. 2016;188:E254–60.
Brunelli L, Jenkins SM, Gudgeon JM, Bleyl SB, Miller CE, Tvrdik T, et al. Targeted gene panel sequencing for the rapid diagnosis of acutely ill infants. Mol Genet Genom Med. 2019;7:e00796.
Diemen CC, van, Kerstjens-Frederikse WS, Bergman KA, Koning TJ, de, Sikkema-Raddatz B, Velde JKvander, et al. Rapid targeted genomics in critically ill newborns. Pediatrics. 2017;140:e20162854.
Kernohan KD, Hartley T, Naumenko S, Armour CM, Graham GE, Nikkel SM, et al. Diagnostic clarity of exome sequencing following negative comprehensive panel testing in the neonatal intensive care unit. Am J Med Genet A. 2018;176:1688–91.
Elliott AM, du Souich C, Lehman A, Guella I, Evans DM, Candido T, et al. RAPIDOMICS: rapid genome-wide sequencing in a neonatal intensive care unit-successes and challenges. Eur J Pediatr. 2019;178:1207–18.
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438.
Powis Z, Hagman KDF, Speare V, Cain T, Blanco K, Mowlavi LS, et al. Exome sequencing in neonates: diagnostic rates, characteristics, and time to diagnosis. Genet Med. 2018;20:1468–71.
Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6.
Kingsmore SF, Cakici JA, Clark MM, Gaughran M, Feddock M, Batalov S, et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105:719–33.
LaDuca H, Farwell KD, Vuong H, Lu H-M, Mu W, Shahmirzadi L, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PloS ONE. 2017;12:e0170843.
Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112:5473–8.
Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med. 2018;3:6.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55.
Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135.
Acknowledgements
The authors would like to thank Lisa Salz, LCGC, and Mary Willis, MD, PhD, for their critical edits regarding pretest counseling.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing and review of this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Carroll, J., Wigby, K. & Murray, S. Genetic testing strategies in the newborn. J Perinatol 40, 1007–1016 (2020). https://doi.org/10.1038/s41372-020-0697-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41372-020-0697-y
This article is cited by
-
Identifying the essential knowledge and skills for Neonatal-Perinatal Medicine: a systematic analysis of practice
Journal of Perinatology (2022)