Abstract
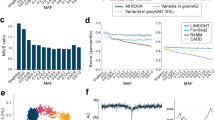
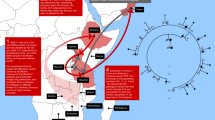
The Fabry disease-causing mutation, the GLA IVS4+919G>A (designated GLA IVS4), is very prevalent in patients with hypertrophic cardiomyopathy in Taiwan. This X-linked mutation has also been found in patients in Kyushu, Japan and Southeast Asia. To investigate the age and the possible ancestral origin of this mutation, a total of 33 male patients with the GLA IVS4+919G>A mutation, born in Taiwan, Japan, Singapore, Malaysia, Vietnam, and the Fujian and Guangdong provinces of China, were studied. Peripheral bloods were collected, and the Ilumina Infinium CoreExome-24 microarray was used for dense genotyping. A mutation-carrying haplotype was discovered which was shared by all 33 patients. This haplotype does not exist in 15 healthy persons without the mutation. Rather, a wide diversity of haplotypes was found in the vicinity of the mutation site, supporting the existence of a single founder of the GLA IVS4 mutation. The age of the founder mutation was estimated by the lengths of the mutation-carrying haplotypes based on the linkage-disequilibrium decay theory. The first, second, and third quartile of the age estimates are 800.7, 922.6, and 1068.4 years, respectively. We concluded that the GLA IVS4+919G>A mutation originated from a single mutational event that occurred in a Chinese chromosome more than 800 years ago.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Desnick RJ. Fabry disease (α-galactosidase A deficiency). In: Encyclopedia of genetics. Berkeley: Elsevier; 2001. p. 681.
Germain DP. Fabry disease. Orphanet J Rare Dis. 2010;5:30. https://doi.org/10.1186/1750-1172-5-30.
Desnick RJ. Fabry disease, an under-recognized multisystemic disorder: expert recommendations for diagnosis, management, and enzyme replacement therapy. Ann Intern Med. 2003;138:338. https://doi.org/10.7326/0003-4819-138-4-200302180-00014.
Nakao S, Kodama C, Takenaka T, Tanaka A, Yasumoto Y, Yoshida A, et al. Fabry disease: detection of undiagnosed hemodialysis patients and identification of a “renal variant” phenotype1. Kidney Int. 2003;64:801–7. https://doi.org/10.1046/j.1523-1755.2003.00160.x.
Nakao S, Takenaka T, Maeda M, Kodama C, Tanaka A, Tahara M, et al. An atypical variant of Fabry’s disease in men with left ventricular hypertrophy. N Engl J Med. 1995;333:288–93. https://doi.org/10.1056/nejm199508033330504.
Wv Scheidt, Eng CM, Fitzmaurice TF, Erdmann E, Hübner G, EGJ Olsen, et al. An atypical variant of Fabry’s disease with manifestations confined to the myocardium. N Engl J Med. 1991;324:395–9. https://doi.org/10.1056/nejm199102073240607.
Desnick RJ, Brady RO. Fabry disease in childhood. J Pediatrics. 2004;144:S20–S6. https://doi.org/10.1016/j.jpeds.2004.01.051.
Lin H-Y, Chong K-W, Hsu J-H, Yu H-C, Shih C-C, Huang C-H, et al. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ Cardiovasc Genet. 2009;2:450–6. https://doi.org/10.1161/circgenetics.109.862920.
Hsu T-R, Hung S-C, Chang F-P, Yu W-C, Sung S-H, Hsu C-L, et al. Later onset Fabry disease, cardiac damage progress in silence. J Am Coll Cardiol. 2016;68:2554–63. https://doi.org/10.1016/j.jacc.2016.09.943.
Ishii S, Nakao S, Minamikawa-Tachino R, Desnick RJ, Fan J-Q. Alternative splicing in the α-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am J Hum Genet. 2002;70:994–1002. https://doi.org/10.1086/339431.
Chang W-H, Niu D-M, Lu C-Y, Lin S-Y, Liu T-C, Chang J-G. Modulation the alternative splicing of GLA (IVS4+919G>A) in Fabry disease. PLOS One. 2017;12:e0175929. https://doi.org/10.1371/journal.pone.0175929.
Lin H-Y, Liu H-C, Huang Y-H, Liao H-C, Hsu T-R, Shen C-I, et al. Effects of enzyme replacement therapy for cardiac-type Fabry patients with a Chinese hotspot late-onset Fabry mutation (IVS4+919G>A). BMJ Open. 2013;3:e003146. https://doi.org/10.1136/bmjopen-2013-003146.
Niu D-M, Yu W-C, Hsu T-R, Chang F-P, Sung S-H, Chu T-H. When is the best time to start enzyme replacement therapy in patients with cardiac-type Fabry disease? Experience from Taiwan, an area highly prevalent in this cardiac phenotype. Mol Genet Metab. 2015;114:S87–S8. https://doi.org/10.1016/j.ymgme.2014.12.194.
Gandolfo LC, Bahlo M, Speed TP. Dating rare mutations from small samples with dense marker data. Genetics. 2014;197:1315–27. https://doi.org/10.1534/genetics.114.164616.
Efron B. Bootstrap methods: another look at the Jackknife, Springer series in statistics. New York: Springer; 1992. p. 569–93.
Carriço JA, Crochemore M, Francisco AP, Pissis SP, Ribeiro-Gonçalves B, Vaz C. Fast phylogenetic inference from typing data. Algorithms Mol Biol. 2018;13. https://doi.org/10.1186/s13015-017-0119-7.
Altshuler D, Gibbs R, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8.
Kanzawa-Kiriyama H, Jinam TA, Kawai Y, Sato T, Hosomichi K, Tajima A, et al. Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan. Anthropological Sci. 2019;127:83–108. https://doi.org/10.1537/ase.190415.
Choi J-H, Lee BH, Heo SH, Kim G-H, Kim Y-M, Kim D-S, et al. Clinical characteristics and mutation spectrum of GLA in Korean patients with Fabry disease by a nationwide survey. Medicine. 2017;96:e7387. https://doi.org/10.1097/md.0000000000007387.
Tadaka S, Katsuoka F, Ueki M, Kojima K, Makino S, Saito S, et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6. https://doi.org/10.1038/s41439-019-0059-5.
Dreyer EL. Japanese piracy in Ming China during the 16th Century. By Kwan-Wai So. East Lansing: Michigan State University Press, 1975. x, 254 pp. J Asian Stud. 1976;36:129–30. https://doi.org/10.1017/s002191180007902x.
Acknowledgements
The authors would like to thank Wen-Ya Ko for his professional advices in population genetics, as well as the journal reviewer #2 for the comprehensive guidance in the presentation of the manuscript.
Funding
This work was partly supported by the Ministry of Science and Technology, Taiwan (grant number 108-2321-B-075-003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Liang, KH., Lu, YH., Niu, CW. et al. The Fabry disease-causing mutation, GLA IVS4+919G>A, originated in Mainland China more than 800 years ago. J Hum Genet 65, 619–625 (2020). https://doi.org/10.1038/s10038-020-0745-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0745-7