Abstract
Paternal inheritance of both Y chromosome and surnames makes it possible to trace the origin and migration histories of surnames based on high-resolution Y chromosome phylogeny. In this study, 292 male samples with surname Ye (叶) in China were collected to unravel the history of this surname. Among these samples, O-F492 showed the highest frequency (26.71%). Analysis based on Y chromosome genotyping data of 52,798 males from virtually the whole China revealed a close correlation between O-F492 and surname Ye. High-throughput sequencing of 131 unrelated male individuals covering all sub-haplogroups in O-F492 was conducted to update the phylogeny of O-F492. Most of the Ye individuals (43/64, 67.19%) are embedded in three major branches, i.e., O-MF1461, O-MF15219, and O-FGC66159, deriving from the same node (O-FGC66168). These three clades restrictively distributed in different regions, likely attributed to independent differentiations. Coalescent ages of the three subclades are estimated ranging from 1,925 to 1,775 years ago, probably driven by the massive migration from north to south China after Yongjia riot in Jin Dynasty, consistent with the migration history of surname Ye. Our study thus shed important light on the history of the surname Ye from genetic perspective.
Similar content being viewed by others
Introduction
The non-recombination region of human Y chromosome (NRY) is strictly inherited paternally from father to son. Similarly, Chinese surnames are passed from father to children in traditional culture, especially in the Han ethnic group [1]. Therefore, male individuals sharing the same surname are expected to possess similar Y chromosomes [2]. This close relationship between Y chromosome and surnames makes Y chromosome to be the optimal material to trace the origin and dispersal histories of surnames.
Well-defined Y chromosome phylogeny based on markers in NRY supplies a widely informative tool to reconstruct the genetic relationship of human populations and paternal lineages, thus making it possible to trace the origin and the migration history of modern humans [3,4,5]. During the last decades, genetic histories of some paternal families had been revealed based on NRY information, including the population expansion in Genghis Khan’s (成吉思汗) case [6, 7], as well as the verification of the descendants of the famous Chinese Emperor CaoCao (曹操) by combining the stemma records [8, 9]. However, few genetic efforts had been carried out to investigate the origin and expansion histories of surnames in China.
In this study, we aimed to explore the origin and migration history of surname Ye (叶) in China based on high-resolution genotyping and sequencing data of Y chromosome. As the 49th most common surname in China according to the sixth national census, the Ye surname distributed mainly in Guangdong, Zhejiang, and Fujian provinces etc. Some historical records suggest that the surname Ye originated in Henan province with their ancestor Ye Gong (叶公) [10, 11]. This Ye Gong would have evolved from surname Shen (沈) whose ancestors form the noble family of Mi (芈) of the Chu kingdom (1115–223 B.C.) [10,11,12]. However, until nowadays, whether the genetic histories of males with this surname match well with the historical records needs further investigation.
To infer the history of surname Ye, we collected saliva samples from 292 unrelated male individuals with surname Ye from China, who are all Han Chinese (Table 1). We first explored the paternal genetic structure of these Ye samples based on genotyping data. Then, the most-common haplogroups of Ye samples were selected to conduct the high-throughput sequencing to update the phylogenetic tree and infer the history of this surname.
Materials and methods
Samples
Two hundred and ninety two unrelated Ye samples were selected from the customer base of Chengdu 23MoFang, Inc., a consumer personal genetics scientific company. This study was conducted in accordance with the human and ethical research principles of the Ministry of Science and Technology of the People’s Republic of China (Interim Measures for the Administration of Human Genetic Resources, 10 June 1998). Informed consents were obtained from all participants under the protocol approved by the Ethical Committee of 23MoFang, Inc. All participants selected in this study provided the detailed ancestral information including both surname and native place.
Y-Chromosome markers and genotyping
Genomic DNA was extracted from the saliva samples, and then genotyped on the Affymetrix genotyping platform of AffyPipe [13], using 23MoFang v1.0 and v2.0 high-density SNP arrays, which included Y chromosome markers ~26,000 and 33,000 SNPs, respectively. Quality control was performed in PLINK V1.07 [14] and the individuals and SNPs with genotype call rate of < 98.5% were excluded. The individuals whose sample analyses failed were recontacted by 23MoFang customer service to provide additional samples, as is done for all 23MoFang customers. Here, we follow the rules defined by Y Chromosome Consortium [15] to update phylogenetic trees of Y chromosome haplogroups.
Targeted capture and library preparation
Genomic DNA of the selected samples were sheared using Bioruptor Pico B01060001 (Diagenode, Belgium) to 150–250 bp length, and then were fixed to blunt-end, added 3’-A tail, and ligated with barcode-linked Illumina paired-end adaptors. Ligation products were amplified by PCR, and 300–350 bp sections were extracted through AgencourtAMPure XP. Then we used the designed library that covers 9.99 million sites of the NRY to enrich the target region [16]. After another round of amplification, the captured products were quantified with the Qubit dsDNA HS Assay Kit (Invitrogen, USA). Paired-end sequencing, which reads 150 bases from each end of the fragment for targeted libraries, was performed using Illumina Novaseq 6000 (Illumina, San Diego, CA). False-positive rate was tested in each sample by calculating the SNP concordance between genotyping and NGS data (Table S10), indicating a low false-positive rate in the genotyping data.
Processing of next-generation sequencing data
A total of 131 unrelated samples (Table S1) covering all sub-haplogroups in O1a1a1a1a1a1-F492 (see results) were selected to conduct high-throughput sequencing in order to update the phylogenetic tree of this haplogroup. These 131 samples included 64 unrelated Ye individuals, as well as 67 individuals with other surnames, such as Zhong (钟), Hong (洪), and Qian (钱), etc. The barcodes were removed and the reads were assigned to each sample with fastp [17]. For paired-end sequencing, the reads were assigned to the same sample only when the both barcodes were identical. The reads were mapped to hg19 using bwa (version 0.5.8) aligner [18], and sam files were generated. Reads that were uniquely mapped on Y chromosome were extracted and transformed into bam file with samtools (version 0.1.8) [19]. Duplication reads were removed by Picard’s MarkDuplicate (http://picard.sourceforge.net) (for paired-end). Indels were re-aligned using GATK [20], following which samtools mpileup and variations were called with the following criteria: for one sample, the position where the alternative allele (compared with hg19) must be ≥2 × coverage and at the same time ≥1/2 of total coverage. All the variance candidates were collected, and genotypes were called on all the sequenced samples. Out of those candidates, SNPs were semi-manually filtered considering consistency to the Y chromosomal phylogeny, coverage (especially for the private SNPs, a minimum of ≥2 × and the mapping quality ≥20) was required.
Time estimation of the nodes in the phylogenetic tree
We use the actual number of mutations (NSNP) to estimate the time to the most recent common ancestor (TMRCA) [21], which is defined as:
The size B of the measured and mapped area of NRY is evaluated using the stably performance sites (8.47 million sites, Table S2) of designed library position. The µ is the per-generation mutation rate with the most common value of NRY ~ 0.82 × 10−9 and 0.76 × 10−9 bp−1 per year. A generation time of 30 years was adopted to convert per-generation rates to yearly rates.
Results
Enrichment of haplogroup O-F492 in Ye samples
Genotyping results indicated that the 292 unrelated Ye individuals can be allocated to 101 different haplogroups, e.g., O1a1a1a1a1a1-F492, O2a2b1a1a-M133, and O2a1c1a1a1a-F11 etc. Specifically, haplogroup O-F492 accounted 26.71% of the Ye samples, significantly higher than other clades (Table S3). Moreover, this haplogroup is shared by Ye samples from different provinces (Table 1), likely represented the common genetic component of males with this surname.
We then pay special attention to haplogroup O-F492. We collected genotyping data of 3,048 male individuals belonging to O-F492 from 52,798 unrelated male samples from virtually the whole China (unpublished data from 23Mongfang). Interestingly, these O-F492 individuals distributed primarily in southern provinces of China, especially in the territory of Low Yangtze River Valley (Jiangsu (10.48%) and Zhejiang (11.57%) provinces) and Guangdong province (9.55%) (Table 1 and Fig. 1a). Of note, this geographic distribution matches well with the distribution of surname Ye (Table 1 and Fig. 1b), while the other surnames distributed different with O-F492 (Table S9 and Fig. S1), thus indicating the close relationship between O-F492 and surname Ye. This correlation finds further supports from the most-significant level of O-F492 in surname Ye (p = 3.53E-30; Table 2 and Table S4). Therefore, this haplogroup can be considered as a potential genetic marker of surname Ye, thus would shed important light on the origin and migration history of this surname.
Geographical distributions of a haplogroup O-F492 and b surname Ye in China. a The percentage of the male individuals belonging to the haplotype O-F492 in every province of China after adjusted based on the sixth national census of China. b The percentage of the male individuals with surname of Ye in every province of China after adjusted based on the sixth national census of China
Updating phylogenetic structure of O-F492 based on sequencing data
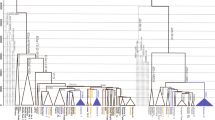
To update the phylogenetic tree of haplogroup O-F492, 131 unrelated O-F492 samples (64 surname Ye and 67 other surnames) (Table S1), were selected to perform high-throughput sequencing (average depth: 130× ; Table S5). A total of 236 SNPs (Table S6) defining (sub-)haplogroups had been identified, within which 157 (started with “MF”) are novel SNPs that had not been reported in previous studies. The updated phylogenetic tree of this haplogroup was shown in Fig. 2 and Table S7, harboring six subclades, including O1a1a1a1a1a1a-F656, O1a1a1a1a1a1b-FGC66168, O1a1a1a1a1a1c-Y31266, O1a1a1a1a1a1d-A12442, O1a1a1a1a1a1e-MF1071, and one newly defined haplogroup, which was tentatively named as O1a1a1a1a1a1f-MF19600. Within haplogroup O1a1a1a1alale, which was defined by MF1071 (8107855, T - > A), MF1072 (16475547, G - > A) and MF1073 (17865165, C - > T) in ISOGG (https://isogg.org/tree/2017/ISOGG_HapgrpO17.html), one sample (YQ0023) shows positive at positions MF1071 (8107855: T - > A, ref/alt: 0/80) and MF1073 (17865165: C - > T, ref/alt: 0/8) and lacks the mutation at MF1072 (16475547: G - > A, ref/alt: 73/0). This indicates that the mutations at MF1071 and MF1073 should be ancestral to the entire haplogroup, whereas the mutational event at MF1072 occurred later. We therefore defined this haplogroup by MF1071 and MF1073 in this study.
The phylogenetic tree of haplotype O-F492. Different branches are shown with different colors. Yellow: O-MF19600; Green: O-MF1071; Purple: O-Y31266; Orange: O-A12442; Turquoise: O-FGC66168; Gray: O-F492; Blueberry: O-MF14611; Aqua blue: O-MF15219; Sky blue: O-FGC66159. The Ye samples are colored by red
Of note is that, the subclades of O-F492 displayed surname-clustering pattern, with different branches restrictively distributed in different surnames (Fig. 2). For example, surname Zhong (钟) individuals are mainly found belonging to O1a1a1a1a1a1c1-Y31261, whereas samples with surname Xin (忻) are identified belonging to haplogroup O1a1a1a1a1a1d1b-MF19468. Similarly, surnames Hong (洪), Qian (钱), and Qu (璩) distributed mainly in O-Y137090, O-MF6069, and O-MF2651, respectively, all of which derived from O-F656. Specifically, the majority (43/64, 67.19%) of Ye samples were found in O1a1a1a1a1a1b1-Z23494, a major subclade of O-FGC66168. Among the six subclades of O-Z23494, three lineages were mainly occupied by surname Ye individuals, including O-MF1461, O-MF15219, and O-FGC66159. Interestingly, these three clades displayed geographic specific distributions, with O-MF1461 and O-MF15219 mainly found in Zhejiang and Jiangsu provinces, whereas O-FGC66159 distributed primarily in Guangdong province, concordant with the geographic distribution of surname Ye. It is thus probable that these branches differentiated independently in different areas after their derivation from O-FGC66168.
The TMRCA of haplogroup O-F492
The private SNPs (Table S8) of each sample and the branch definition SNPs were used to calculate the TMRCA (Table S11). Results indicated that haplogroup O-F492 is relatively young with a divergence time of 2,950 years ago (ya). Similarly, the coalescent ages of its sub-haplogroups, including O-F656, O-FGC66168, O-Y31266, O-A12442, O-MF1071, and O-MF19600, were estimated ranging from 2,075 to 2,950 ya. This implied that subsequent expansions of these sublineages after their differentiations from the ancestor node, O-F492. The three major branches that are specific in surname Ye, i.e., O-MF14611, O-MF15219, and O-FGC66159, are coalesced to 1,775 ya, 1,925 ya, and 1,825 ya, respectively. These results demonstrated at least two expansions of haplogroup O-F492 during the historical period, which probably related to expansions of males with surname Ye.
Discussion
In this study, we found an enrichment of haplogroup O-F492 in surname Ye samples (26.71%). Large-scale data set from virtually the whole China further confirmed a close correlation between O-F492 and surname Ye. Based on updated phylogeny of O-F492, we identified a star-like phylogenetic structure of this haplogroup, likely attributed to a rapid population expansion at ~2,950 ya. In fact, this timeframe overlapped with the Western Zhou Dynasty (c. 11th century-771 B.C.), during which the first massive southward migration of Ye family occurred [12]. It is therefore probable that the population expansion during Western Zhou Dynasty triggered the differentiation and the first migration of surname Ye, as well as other surnames, such as Zhong, Hong and Qian etc., which also occupied specific clades in O-F492.
Specifically, one of the major sub-branches of O-F492, O-FGC66168 (especially its major subclade O-Z23494), was found specific in our Ye samples. Interestingly, the only one root type of O-FGC66168 was from Henan province in northern China (Fig. 2), indicating its potential northern China origin. Three sub-haplogroups of the O-Z23494, viz., O-MF14611, O-MF15219, and O-FGC66159, showing star-like phylogenetic structures, would reflect population expansions of males with surname Ye during 1,925 to 1,775 ya. This timeframe matches well with the period after the Yongjia chaos in 311 A.D. in Jin Dynasty, which caused the first one of the three massive migrations from north to south in Chinese history. It is therefore possible that the males with surname Ye had also been involved into this southward migration from northern China, consistent with second southward migration of Ye family according to historical records [10, 12]. Given their different geographic distributions, these three clades would have differentiated independently in separate areas, likely through founder effects.
Taken together, our study revealed that Y chromosome haplogroup O-F492 has close relationship with migrations of surname Ye, and will shed important light on the origin and migration of this surname. However, one should be cautious of the sharing of O-F492 between Ye and other surnames, as well as the existence of other haplogroups (e.g., O-M133) in Ye individuals. Therefore, it is also probable that surname changes and multiple origins had also occurred during the formation history of this surname. In addition, we only explored individuals of surname Ye in Han Chinese, the Ye individuals from other ethnic groups in China, however, had not been considered in this study, making the possibility that surname Ye evolved from the southern Chinese ethnic groups [10, 11] uncertified. Moreover, besides surname Ye, whether the match between surnames and Y haplogroups is common in other Chinese surnames needs further investigations. More studies based on large-scale samples and high-resolution Y chromosome data set are needed to intensively unravel the formation history of surname Ye, as well as the other surnames.
References
Wang CC, Li H. Inferring human history in East Asia from Y chromosomes. Invest Genet. 2013;4:11.
Sole-Morata N, Bertranpetit J, Comas D, Calafell F. Y-chromosome diversity in Catalan surname samples: insights into surname origin and frequency. Eur J Hum Genet. 2015;23:1549–57.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–8.
Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA. 2009;106:20174–9.
Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, et al. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 2005;77:408–19.
Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, et al. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–21.
Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, et al. Recent spread of a Y-chromosomal lineage in northern China and Mongolia. Am J Hum Genet. 2005;77:1112–6.
Wang C, Yan S, Hou Z, Fu W, Xiong M, Han S, et al. Present Y chromosomes reveal the ancestry of Emperor CAO Cao of 1800 years ago. J Hum Genet. 2012;57:216–8.
Wang CC, Yan S, Yao C, Huang XY, Ao X, Wang Z, et al. Ancient DNA of Emperor CAO Cao’s granduncle matches those of his present descendants: a commentary on present Y chromosomes reveal the ancestry of Emperor CAO Cao of 1800 years ago. J Hum Genet. 2013;58:238–39.
Ye Hongying. Historical exploration of surname Ye. Science and Technology Innovation Guide; 2007. p. 226–226.
People’s Government of Ye County. Tracing the source of Surname Ye: Ye Gong and Ye County Research Collection. Zhongzhou: Ancient Books Publishing House; 2000.
Wu Jianhua. The Origin of Huizhou Ye Surname Based on the Relationships between Regional Surname Research and Chinese Surnames, Clan History, and Genealogy. Chinese Social History Review; 2009. p. 134–149.
Nicolazzi EL, Iamartino D, Williams JL. AffyPipe: an open-source pipeline for Affymetrix Axiom genotyping workflow. Bioinformatics. 2014;30:3118–19.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–48.
Poznik GD, Henn BM, Yee MC, Sliwerska E, Euskirchen GM, Lin AA, et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science. 2013;341:562–65.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bionformatics. 2018;34:i884–i890.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Adamov D, Guryanov V, Karzhavin S, Tagankin V, Urasin V. Defining a new rate constant for Y-chromosome SNPs based on Fullsequencing data. Russ J Genet Geneal. 2015;1:68–89.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31620103907, 31601017), Strategic Priority Research Program (Grant No. XDA20040102), Chinese Academy of Sciences (QYZDB-SSW-SMC020), Yunnan Applied Basic Research Project (2017FB044), and CAS “Light of West China” Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zeng, Z., Tian, J., Jiang, C. et al. Inferring the history of surname Ye based on Y chromosome high-resolution genotyping and sequencing data. J Hum Genet 64, 703–709 (2019). https://doi.org/10.1038/s10038-019-0616-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0616-2