Abstract
Intellectual disability (ID) is a genetically heterogeneous neurodevelopmental disorder characterised by significantly impaired intellectual and adaptive functioning. ID is commonly syndromic and associated with developmental, metabolic and/or neurological findings. Autosomal recessive ID (ARID) is a significant component of ID especially in the presence of parental consanguinity. Several ultra rare ARID associated variants in numerous genes specific almost to single families have been identified by unbiased next generation sequencing technologies. However, most of these new candidate ARID genes have not been replicated in new families due to the rarity of associated alleles in this highly heterogeneous condition. To determine the genetic component of ARID in a consanguineous family from Turkey, we have performed SNP-based linkage analysis in the family along with whole exome sequencing (WES) in an affected sibling. Eventually, we have identified a novel pathogenic variant in EEF1D, which has recently been recognised as a novel candidate gene for ARID in a single family. EEF1D encodes a ubiquitously expressed translational elongation factor functioning in the cytoplasm. Herein, we suggest that the loss of function variants exclusively targeting the long EEF1D isoform may explicate the ARID phenotype through the heat shock response pathway, rather than interfering with the canonical translational elongation.
Similar content being viewed by others
Introduction
Intellectual disability (ID) comprises a group of neurodevelopmental disorders, which are basically characterised by significant limitations in both intellectual functioning and adaptive behaviours starting before the age of 18 [1]. ID associated with mild to severe learning and behavioural defects may be the only apparent manifestation of the disease (nonsyndromic ID) or more commonly, it may be accompanied by congenital malformations, neurological or metabolic findings (syndromic ID) [2]. Therefore, ID is an extremely heterogeneous condition that causes cognitive impairment due to both genetic and non-genetic factors. Well-established genetic causes of ID include chromosomal anomalies and monogenic diseases, while the environmental factors that may contribute to ID include foetal alcohol exposure, infections, malnutrition and insufficient health care [3]. In populations where parental consanguinity rate is high, ID is assumed to be inherited mostly as autosomal recessive [1]. Accordingly, autosomal recessive ID (ARID) is mostly caused by private mutations in families due to extreme genetic heterogeneity, which can be resolved by unbiased whole genome approaches [3, 4].
Herein, we report three sisters with ARID born to consanguineous parents from Turkey presenting with similar accompanying clinical features. Using whole genome SNP genotyping followed by linkage and whole exome sequencing (WES) analyses, we have identified a novel pathogenic variant in EEF1D, which has recently been proposed as a candidate gene for ARID in a single family [4]. We propose that biallelic loss of function variations of EEF1D in these two families affect only the heat-shock response (HSR) pathway triggered by alternatively spliced long isoform of EEF1D.
Materials and methods
Patients and clinical assessments
A family from Turkey with first cousin parents (E11-203 and E11-204), three affected girls (E11-205, E11-207 and E11-206) and two unaffected boys (E12-45 and E12-46) has been clinically analysed at Harran University, Faculty of Medicine, Department of Paediatric Neurology, Sanliurfa, Turkey (Fig. 1). Physical and neurological examinations were performed for all available family members and detailed information on family history was collected. Magnetic resonance imaging (MRI) of the brain and electroencephalography (EEG) recordings were performed on the affected siblings, whenever possible. Informed consents were obtained from all family members and control individuals in accordance with Istanbul University, Istanbul Faculty of Medicine, Clinical Ethics Committee. DNA was extracted from peripheral blood samples of both children and their parents using QIAamp DNA Blood Maxi Kit (Qiagen GmbH, Hilden, Germany).
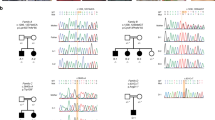
Genetic studies in the ARID family. a Multipoint LOD scores (Allegro) of the total SNP data set along the autosomes. b SNP derived haplotypes on chromosome 8. Haplotypes of the parents and siblings have been shown in side-by-side alignment in order the track the IBD segregation. c Segregation of NM_001130053.3 (ENST00000423316.6):c.948G>A variant within the family. d Schematic representation of selected EEF1D transcripts ENST00000317198.10 and ENST00000423316.6 encoding the eEF1Bδ1 and eEF1BδL isoforms, respectively. The exon unique to the large isoform and the protein domain encoded by this exon are shown in pink. Sites of ARID specific variations in each transcript and corresponding amino acid changes on eEF1BδL isoform are also indicated
Linkage analysis and haplotyping
All seven family members were SNP genotyped using Illumina HumanCytoSNP-12 BeadChip kit and subjected to whole genome logarithm of the odds (LOD) score analysis using ALLEGRO version 1.2c software under software package easyLINKAGE plus version 5.08 assuming recessive inheritance with full penetrance [5]. Haplotypes in regions with positive linkage peaks were manually inspected for identical by descent (IBD) inheritance of the shared haplotypes in the affected children. SNP array data was also used for molecular karyotyping to detect copy number variations (CNVs) using Illumina GenomeStudio version 2011.1 and its plug-in application cnvPartition v3.1.6. CNV calling was performed with a confidence threshold value of 35 and a minimum tandem probe number of 3. Sex chromosomes were also analysed both for CNV detection and sample sex crosscheck. Copy neutral homozygous regions were detected by a default minimum value of 1 Mb.
Exome sequencing and segregation analyses
WES was performed for one of the affected girls (Patient E11-207) through the service provided by Oxford Gene Technology (OGT; Oxford, UK). Exonic DNA was captured with the SureSelect Human All Exon V4 Kit (Agilent Technologies, Santa Clara, CA, USA) and sequenced on the Illumina HiSeq2000 platform. The sample was sequenced to a mean target coverage of 50× with 90.94% of bases covered at a depth of >20×. Exome data analysis was completed in OGT’s pipeline and variant lists were browsed using OGT’s proprietary software (for details [6]). All rare variants in selected linkage intervals with a carrier frequency expected for an ultra rare recessive disorder (alternative allele frequency; AAF < 0.14%) were filtered [4, 7] (Supplementary information). These variants were then re-annotated with ENSEMBL Variant Effector Predictor Tool (VEP) for both detecting the consequence of these variants on current transcript versions and retrieving up-to-date population frequency data. Sequence validation and segregation analyses were performed via Sanger sequencing for each candidate variant using standard procedures. Hundred and eighty four additional individuals from Turkey with no apparent history of neurological disorders were screened via Sanger sequencing for selected variants.
Results
Clinical presentation
We have studied a consanguineous family from Turkey having three affected girls along with two unaffected boys (Fig. 1). The patients E11-205, E11-207 and E11-206 were examined at our paediatric neurology outpatient clinic at Harran University Faculty of Medicine at the ages of 12, 8 and 4, respectively, and enrolled to this study due to global developmental delay, ID and epileptic findings. None of the patients could talk. E11-205, who was the first child of the family, was born full term after uncomplicated pregnancy by normal vaginal delivery. She had microcephaly and facial dysmorphic features including hypertelorism and micrognathia. She had weak head and neck control and she was able to sit, stand and walk only with support possibly due to hypotonia. Her deep tendon reflexes were brisk and bilateral Babinski sign was present. She had a history of generalised convulsions starting from the age of 3. According to her parents, the frequency of the seizures was once in a month, which could not be controlled due to insufficient utilisation of prescribed antiepileptic drugs. The clinical characteristics of her both younger sisters were almost identical. These two patients were available for MRI and EEG analyses. In both patients, MRI was indicative of thin corpus callosum and interictal EEG showed generalised epileptiform discharges originating from the left frontotemporal area and spreading to both brain hemispheres. At the time of examination, Patient E11-206 was seizure free for almost one year, unlike her older affected sisters.
SNP Array, WES and segregation analyses
CNV analysis of the SNP data did not reveal any pathogenic CNVs shared in the affected individuals. Parametric linkage analysis in the family revealed two linkage peaks on chromosomes 8 and 9 with maximum LOD scores of 2.65 and 2.5, respectively (Fig. 1). Haplotype inspection in these loci revealed segregation of IBD haplotypes consistent with an autosomal recessive mode of inheritance only for the distal portion of chromosome 8 (8q24-qter). Region on chromosome 9q21.33–34.12 was eliminated as the unaffected mother was found to be homozygous between rs17088766 and rs11244140 (hg38; chr9:85,539,994–130,787,671 bp) throughout the locus encompassing the positive LOD score peaks. The linkage interval at the distal portion of chromosome 8 was marked by informative rs6578128 and contained 173 genes as listed by NCBI Genome Data Viewer (hg38; chr8:140,642,366–145,138,636 bp). WES data of E11–207 was filtered for this interval on chromosome 8q24-qter. Table 1 summarises all three variants identified via exome sequencing in E11–207, which were further validated by Sanger sequencing and segregation analysis. All three variants were absent from the reference cohort screened from Turkey. Amongst, EEF1D was selected as the most likely gene associated with the phenotype in the family as it had previously been implicated in ARID [4].
Discussion
Although, ID is collectively a common disorder with a prevalence approaching to 1% globally, almost each ID subtype comprises a rare disorder, bringing out its own needs for effective diagnosis, counselling and treatment options [8]. ARID represents a significant portion of ID, especially in populations with high rate of parental consanguinity. Unbiased genomic approaches in large numbers of ARID families have identified novel candidate genes, but pathogenicity has not been replicated for most of the ‘private genes’ with a variation segregating only in a single family [4, 9]. One such gene; EEF1D has recently been identified as an ARID candidate in a consanguineous family from Syria with three affected children afflicted with severe ID, microcephaly and short stature in a collaborative exome sequencing effort of 152 consanguineous families with neurodevelopmental disorders [4]. The findings presented herein add support for EEF1D as an important gene for ARID by identifying a novel EEF1D variation in a second family from Turkey with ID and developmental delay.
EEF1D can be described as a multipurpose gene in the sense that it gives rise to at least four protein isoforms in humans (ENSEMBL Gene ID: ENSG00000104529). These are grouped as short and long isoforms, where there are three short isoforms (eEF1Bδ1–3) each with 281, 262 or 257 amino acids, respectively and one long isoform (eEF1BδL) with 647 amino acids [10]. A single, but large coding exon that is alternatively spliced out in the transcripts of short isoforms, but retained in the long ones is responsible for this drastic difference between the lengths of encoded polypeptides. The canonical eEF1Bδ isoform acts as a translational elongation factor in the cytoplasm by integrating into the multi-subunit eEF1 complex [11]. The long isoform eEF1BδL is translocated to the nucleus through its N-terminus nuclear localisation signal (NLS) sequence and act as a HSR transcription factor [12]. The two EEF1D variants presented herein and in the original study both target specifically the large exon retained only in the EEF1D transcripts encoding eEF1BδL isoform (Table 2). This in turn may create an apparent knock-out effect for eEF1BδL function possibly due to translation dependent nonsense-mediated decay of these particular transcripts. The exclusive locations of these two variations are possibly not affecting the transcripts encoding eEF1Bδ, due to their relative deep intronic localisations (Table 2).
The selective eEF1BδL knockout scenario possibly mimicking the effects of two EEF1D variations discussed herein has been studied extensively by the group that has first characterised this isoform [12]. They have shown that selective inhibition of eEF1BδL expression affects cell viability under thermal stress [12]. Very recently, this group has generated an eEF1BδL knock-out mouse model lacking the large EEF1D exon [13]. Interestingly, these mice developed normal until being subjected to fear conditioning tests. After these tests, their brains were found to be decreased in weight with atrophic features in hippocampus and midbrain and reduced cortical layer thickness. Some also developed audiogenic seizures. These findings suggested an external stimuli dependent role for eEF1BδL in brain function.
The dual function of EEF1D in translation elongation and HSR pathway makes it a key element of proteostasis, by which protein homoeostasis is sustained against altering physicochemical conditions and cellular stress factors [14]. Several cellular stress response pathways including HSR assist proteostasis network via spatial-temporal control of molecular chaperons, translational control and degradation of misfolded proteins, which are crucial to cellular function and survival [15]. Stress induced increase in expression of eEF1BδL and decrease of eEF1Bδ will in turn activate HSR pathway and attenuate translation in general [10]. Accordingly, homozygous loss of function variations exclusive for eEF1BδL will alter proper activation of the HSR pathway, while leaving the eEF1Bδ function intact. Consequently, eEF1BδL may be an important stress-related transcription factor that can bind to HSE and HSE-like cis-elements and modulate several cellular stress responses including heat shock and oxidative stress responses.
Regulation of stress responses is a vital process for organisms, as major neurodegenerative diseases including Alzheimer’s, Parkinson’s and Huntington’s diseases as well as amyotrophic lateral sclerosis are characterised by protein misfolding and aggregation; a significant defect in proteostatis [16]. Additionally, several genes encoding members of heat shock stress interactome including METTL23 (methyltransferase like 23), SACS (sacsin molecular chaperon), DNAJC6 (DnaJ heat shock protein family (Hsp40) member C6) and DNAJC12 (DnaJ heat shock protein family (Hsp40) member C12) have been implicated in ID [17,18,19,20,21,22,23].
The same study describing EEF1D has also associated a homozygous splice site variation in TRAP1 (rs1053534094: NM_016292.2:c.1941-1G>A and NM_001272049.1:c.1782-1G>A) with ID [4]. Interestingly, TRAP1 encodes a mitochondrial chaperone protein that is member of the heat shock protein 90 family and has recently been associated with late onset Parkinson’s disease in the homozygous state (rs141984778: NM_016292.2:c.139C>T; p.Arg47* and NM_001272049.1:c.89-1798C>T) [24]. It is very interesting to note that the TRAP1 transcript encoding the large isoform has a 159 bp coding exon that is absent from the short isoform; a similar scenario to EEF1D presented herein. The homozygous stop-gain variant in the late onset Parkinson’s disease case lies within this particular exon leaving the short isoform intact. It is tempting to speculate that a truncating variant affecting both transcripts may lead to a more severe, early onset neurodevelopmental disorder, while a similar variant affecting targeted transcripts may be associated with a late onset neurodegenerative course.
Tissue-specific differences in expression of components of the proteostatis network could serve as the basis for the understanding of the disease pathogenesis in neuronal cells. It has previously been shown that eEF1BδL expression in adult mice on both protein and mRNA levels is restricted to brain and testis, while eEF1Bδ exhibits a more widespread expression pattern [12]. Disruption of the pivotal role of EEF1D during development, where there is dynamic protein synthesis coupled by active chaperone function and rapid stress response may have drastic effects on neurons and lead to severe congenital neurological conditions.
Pathogenic variants in genes encoding eEF1 subunits may cause neurodevelopmental disorders (reviewed in [11]). It seems surprising to have a rather distinct neurological phenotype when such canonical genes are altered. This dilemma may be resolved at least for EEF1D. The two loss of function variations associated with ID reside on a very niche position of the gene and present an isoform and tissue-specific effect [11]. We propose that ID associated variants exert their effect through HSR related transcriptional control rather than canonical translation. Understanding the interplay between alternative mechanisms of proteostatis and the reflection of these delicate pathways on human diseases can bring new insights for better treatment targets. Isoform specific devastating variations are probably an important aspect of disease pathogenesis and may explain the targeted neurological phenotypes in ubiquitously expressed genes.
References
Musante L, Ropers HH. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–9.
van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104.
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatry. 2017;74:293–9.
Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–7.
Abdel-Salam G, Thoenes M, Afifi HH, Körber F, Swan D, Bolz HJ. The supposed tumor suppressor gene WWOX is mutated in an early lethal microcephaly syndrome with epilepsy, growth retardation and retinal degeneration. Orphanet J Rare Dis. 2014;9:12.
Hennekam RC. Care for patients with ultra-rare disorders. Eur J Med Genet. 2011;54:220–4.
Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32:419–36.
Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63.
Kaitsuka T, Matsushita M. Regulation of translation factor EEF1D gene function by alternative splicing. Int J Mol Sci. 2015;16:3970–9.
McLachlan F, Sires AM, Abbott CM. The role of translation elongation factor eEF1 subunits in neurodevelopmental disorders. Hum Mutat. 2018.
Kaitsuka T, Tomizawa K, Matsushita M. Transformation of eEF1Bδ into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011;12:673–81.
Kaitsuka T, Kiyonari H, Shiraishi A, Tomizawa K, Matsushita M. Deletion of long isoform of eukaryotic elongation factor 1Bδ leads to audiogenic seizures and aversive stimulus-induced long-lasting activity suppression in mice. Front Mol Neurosci. 2018;11:358.
Díaz-Villanueva JF, Díaz-Molina R, García-González V. Protein folding and mechanisms of proteostasis. Int J Mol Sci. 2015;16:17193–230.
Weindling E, Bar-Nun S. Sir2 links the unfolded protein response and the heat shock response in a stress response network. Biochem Biophys Res Commun. 2015;457:473–8.
Yerbury JJ, Ooi L, Dillin A, Saunders DN, Hatters DM, Beart PM, et al. Walking the tightrope: proteostasis and neurodegenerative disease. J Neurochem. 2016;137:489–505.
Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210.
Bernkopf M, Webersinke G, Tongsook C, Koyani CN, Rafiq MA, Ayaz M, et al. Disruption of the methyltransferase-like 23 gene METTL23 causes mild autosomal recessive intellectual disability. Hum Mol Genet. 2014;23:4015–23.
Parfitt DA, Michael GJ, Vermeulen EG, Prodromou NV, Webb TR, Gallo JM, et al. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum Mol Genet. 2009;18:1556–65.
Ali Z, Klar J, Jameel M, Khan K, Fatima A, Raininko R, et al. Novel SACS mutations associated with intellectual disability, epilepsy and widespread supratentorial abnormalities. J Neurol Sci. 2016;371:105–11.
Köroğlu Ç, Baysal L, Cetinkaya M, Karasoy H, Tolun A. DNAJC6 is responsible for juvenile parkinsonism with phenotypic variability. Park Relat Disord. 2013;19:320–4.
Gorenberg EL, Chandra SS. The role of co-chaperones in synaptic proteostasis and neurodegenerative disease. Front Neurosci. 2017;11:248.
van Spronsen FJ, Himmelreich N, Rüfenacht V, Shen N, Vliet DV, Al-Owain M, et al. Heterogeneous clinical spectrum of DNAJC12-deficient hyperphenylalaninemia: from attention deficit to severe dystonia and intellectual disability. J Med Genet. 2017.
Fitzgerald JC, Zimprich A, Carvajal Berrio DA, Schindler KM, Maurer B, Schulte C, et al. Metformin reverses TRAP1 mutation-associated alterations in mitochondrial function in Parkinson’s disease. Brain. 2017;140:2444–59.
Acknowledgements
The authors are grateful to the patients and their relatives for their participation in this study. The authors would also like to acknowledge Prof Dr. Akin Iscan for his assistance. This work was supported by the grants of Scientific Research Projects Coordination Unit of Istanbul University, Project Number: 52853 and The Scientific and Technology Research Council of Turkey (TUBITAK) Project Number: 113S331. Biobanking support was given by Istanbul Development Agency (Project Number: TR10/15/YNK/0093). EY and YK have been fellows of TUBITAK Project Number: 113S331. The variants presented in Table 1 have been submitted to ClinVar database (Accession Numbers: SCV000864187, SCV000864186 and SCV000864188).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ugur Iseri, S.A., Yucesan, E., Tuncer, F.N. et al. Biallelic loss of EEF1D function links heat shock response pathway to autosomal recessive intellectual disability. J Hum Genet 64, 421–426 (2019). https://doi.org/10.1038/s10038-019-0570-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0570-z
This article is cited by
-
Two rare autosomal recessive neurological disorders identified by combined genetic approaches in a single consanguineous family with multiple offspring
Neurological Sciences (2024)
-
Re-evaluation and re-analysis of 152 research exomes five years after the initial report reveals clinically relevant changes in 18%
European Journal of Human Genetics (2023)