Abstract
Background
Age-dependent differences in pharmacokinetics exist for metabolically cleared medications. Differential contributions in the cytochrome P450 3A (CYP3A), CYP2C, and flavin-containing monooxygenases (FMOs) families have an important role in the metabolic clearance of a large number of drugs administered to children.
Methods
Unlike previous semiquantitative characterization of age-dependent changes in the expression of genes and proteins (western blot analysis), this study quantifies both gene and absolute protein expression in the same fetal, pediatric, and adult hepatic tissue. Expression was then correlated with the corresponding functional activities in the same samples.
Results
CYP3A and FMO families showed a distinct switch from fetal (CYP3A7 and FMO1) to adult isoforms (CYP3A4 and FMO3) at birth, whereas CYP2C9 showed a linear maturation from birth into adulthood. In contrast, analysis of CYP2C19 revealed higher expression and catalytic efficiency in pediatric samples compared with that in fetal and adult samples. Further, CYP3A and FMO enzymes exhibited an unexpectedly higher functional activity in fetal samples not entirely explained by protein expression.
Conclusion
These surprising findings suggest that CYP and FMO enzymes may encounter development-related differences in their microenvironments that can influence the enzyme activity in addition to protein expression levels.
Similar content being viewed by others
Main
For nearly 75% of the top 200 drugs on the market, hepatic clearance is the predominant clearance mechanism, and over three quarters of hepatic clearance is because of phase I drug-metabolizing enzymes (DMEs) (1). It is recognized that because of ontogeny of DMEs, many drugs exhibit differential pharmacokinetics (PK) in children of different age groups compared with adults. However, our ability to predict PK in children based on this knowledge is somewhat limited because of many factors that are under active investigation. In a study comparing the PK of 45 drugs, children aged 2–12 years had a higher average clearance and lower half-life compared with adults owing to age-related differences in metabolic clearance (2). These PK differences provide a basis for higher weight-based dosing for many drugs (3, 4, 5, 6), including voriconazole, a drug that exhibits nearly threefold higher clearance in children (2–10 years old) than in adults (7). Voriconazole is cleared predominantly by oxidative metabolism through cytochrome P450 (CYP) 3A4, CYP2C19, and flavin-containing monooxygenases (FMO) 3 (refs 8, 9). Early studies examined age-dependent differences in the relative expression of major DMEs responsible for medications cleared by oxidative metabolism, where protein expression was assessed via western blotting or differential metabolism (10, 11, 12, 13). Western blot studies are semiquantitative, and are sometimes limited by difficulties in the selectivity and cross-reactivity of the reagents used, which is overcome by using quantitative LC/MS-based methods to assess protein expression. Of note, a unique trend of higher CYP2C19 expression in pediatric samples (ages <10) over that in the older children or adults was overlooked, leaving unanswered questions about how differences in expression affect the activity of the CYP2C family in different populations (10).
The aim of this study was to quantify cytochrome P450 3A (CYP3A), CYP2C, and FMO gene and absolute protein expression (14, 15, 16) in fetal, pediatric, and adult hepatic tissue, and to correlate the gene and protein expression values with the corresponding functional activities in the same samples. We believe that a higher degree of fidelity achieved using absolute quantitation of enzyme proteins and measuring activity and protein expression in the same samples will enable the use of in vitro metabolism data to explore the knowledge gaps that limit current attempts to determine the factors responsible for observed differences in metabolic clearance for certain drugs among age groups spanning birth to adulthood.
Methods
Chemicals, Reagents, and Sample Selection
Testosterone, 6-β OH-testosterone, S-mephenytoin, 4′-OH S-mephenytoin, diclofenac, 4′-OH diclofenac, cimetidine, NADPH, and voriconazole were purchased from Sigma-Aldrich (St Louis, MO). Cimetidine sulfoxide was purchased from Abcam (Cambridge, MA). Liver tissues from adult (>18 years old, median 39 years), pediatric (aged 1.9 months to 10 years, median 1.5 years), and fetal (15–29 weeks’ gestation, median 16 weeks) donors were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders (Contract #HHSN275200900011C, Ref. No. NO1-HD-9-0011; Baltimore, MD) under an approved UNC-Chapel Hill IRB. Fetal samples were obtained from terminated pregnancies. Pediatric tissues and adult tissues were from healthy donors (e.g., cause of death from trauma or sudden infant death syndrome) or donors with no known hepatic dysfunction that could affect results. Females represented two out of the seven fetal samples, five out of sixteen pediatric samples (with three samples of unknown gender), and six out of ten adult samples. Full donor information is listed in Supplementary Table S1 online.
Preparation of Microsomes
Human liver microsomes (HLMs) were prepared as previously reported (17). Briefly, 0.5 mg of hepatic tissue was homogenized in a buffer containing 50 mM Tris-HCl, 1 mM EDTA, and 154 mM KCl with protease inhibitors (pH 7.4). After homogenization, samples were centrifuged at 10,000g for 30 min, the supernatant was collected, and this step was repeated. The supernatant was mixed with a second buffer containing 100 mM sodium pyrophosphate and 0.1 mM EDTA (pH 7.4) and then centrifuged at 100,000g for 70 min. This step was repeated one final time. The resulting pellet was then resuspended in a buffer containing 10 mM potassium phosphate, 1 mM EDTA, and 20% glycerol (pH 7.4). Microsomal protein concentration was measured using a BCA assay. HLMs were stored at 20 μl aliquots at −80 °C.
Gene and Protein Expression
Liver tissue cells (25 mg) were lysed using Qiazol (Qiagen, Hilden, Germany), sonicated until completely homogenized, and then centrifuged at 10,000g for 12 min. Phase separation and binding of the mRNA was completed using a Qiagen RNEasy Kit. RNA purity and concentration was measured on a NanoDrop 2000 UV–Vis Spectrophotometer (Thermo Scientific; Wilmington, DE) and RNA was diluted to 500 ng/μl for each sample. To ensure sample purity, only samples with an A260/A280 ratio between 1.9 and 2.1 were evaluated for gene expression. The isolated RNA was mixed with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) and incubated at 25 °C × 5 min, 42 °C × 30 min, and then 85 °C × 5 min to synthesize cDNA. The cDNA was mixed with iTaq Supermix (Bio-Rad) and TaqMan Gene Expression Assay for CYP3A4, 3A5, 3A7, 2C9, 2C19, FMO1, FMO3, and 18s (Applied Bio-systems, Foster City, CA). Expression for each enzyme gene was measured (quantitative reverse-transcriptase PCR) and normalized to 18s rRNA. For protein quantitation of CYP3A and CYP2C families (signature peptide sequences shown in Supplementary Table S2 online), HLMs were subjected to liquid chromatography multiple reaction monitoring (LC-MRM)-based targeted quantitative proteomic analysis using recombinant proteins (Supersomes; Corning Gentest, Woburn, MA) of known concentration as calibration standards (14, 16, 18). A more detailed description of the targeted quantitative proteomic methods was previously published (16, 19).
Functional Activity
Six samples from each population (fetal, pediatric, and adult) were chosen to determine differences in functional activity. Samples were selected at random with no prior knowledge about any enzyme activity of the samples. However, our random selection was limited by amount of tissue available. The HLM (0.1 mg/ml) or recombinant enzyme (0.05 mg/ml) along with the probe substrate of each respective enzyme (Table 1) was warned in phosphate buffer (pH 7.4) containing MgCl2 (3 mM) at 37 °C for 5 min. Functional activity reactions were initiated with the addition of NADPH (1 mM) and incubated up to 60 min in triplicate. Aliquots (100 μl) of samples were taken at specified time points, quenched with 300 μl methanol-containing internal standard (0.1 μM of either voriconazole for the CYP assays or phenformin for the FMO assay). Samples were then centrifuged at 10,000g × 10 min and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Metabolite Analysis
The LC-MS/MS system consisted of mobile phase (A) 0.1% formic acid in water and (B) 0.1% formic acid in methanol with a total flow rate of 0.6 ml/min. A Sciex API-4000 triple-quadrupole mass spectrometer was used with a Zorbax SB-C18 column (2.1 × 50 mm, 5 μm). Samples were ionized using an electron spray ionization (ESI) ion source and, following the injection of 10 μl of supernatant, positive ions were monitored at the following MRM Q1/Q3 transitions (m/z): 235.2→150.2 for 4′-OH S-mephenytoin, 312.3→230.2 for 4′-OH diclofenac, and 350.2→127.2 for voriconazole. For the testosterone assay, samples were ionized using an atmospheric pressure chemical ionization (APCI) ion source and monitored at the following MRM Q1/Q3 transitions (m/z): 289.2→97.1 for testosterone, 305.1→269.3 for 6β-OH testosterone, and 350.2→127.2 for voriconazole (16, 20, 21).
Statistical Analysis
Differences between multiple populations were analyzed using Kruskal–Wallis and Dunn’s post hoc test to correct for multiple comparisons and tests between two groups were analyzed using Wilcoxon signed-rank test. The linearity of maturation profiles and correlation analyses were determined by fitting a linear regression curve and 95% confidence intervals to the model. For all analyses, a P value less than 0.05 was considered significant.
Results
CYP3A, CYP2C, and FMO Expression
Relative gene expression trends of the three major CYP3A enzymes (Figure 1a) corresponded well to the trends in protein expression (Figure 2). However, statistical significance for the differences in expression could only be achieved for proteins because of high variability observed in gene expression. As expected, fetal samples had very high expression of CYP3A7 protein (0.535 nmol/mg) with a corresponding low expression of CYP3A4 protein (0.0003 nmol/mg). During maturation, CYP3A7 protein levels decreased in the pediatric and adult populations (0.051 and 0.019 nmol/mg, respectively; Figure 2a). The adult CYP3A7 expression is slightly skewed by a high expression in one sample from an 80-year-old patient, which, if excluded, reduces the adult CYP3A7 expression to 0.0052 nmol/mg. On the other hand, CYP3A4 levels increased from undetectable levels in the fetal period until it reached adult levels (0.069–0.078 nmol/mg) around 1 year of age. Abundance of CYP3A7 was over 10-fold higher and abundance of CYP3A4 was over 200-fold lower in the fetal tissues compared with either the pediatric or adult tissues (P<0.01; Figure 2d). This switch from CYP3A7 as the predominant isoform in fetal tissues to CYP3A4 in adult tissues was seen previously in the literature (13). CYP3A5 remains relatively stable (0.001–0.005 nmol/mg) across all three groups with no statistically significant difference among the three populations. Taken together, gene and protein expression data demonstrate that there is a clear transition of the predominant CYP3A isoform from the fetal population (CYP3A7) into adulthood (CYP3A4). These results generally confirm the pattern of age-related changes in the expression of CYP 3A enzymes that is reported previously (12, 13, 22).
Expression of CYP and FMO genes in fetal, pediatric, and adult hepatic tissues. Gene expression was normalized to the lowest expressed enzyme, specifically adult FMO1, in order to show relative expression of all enzymes in all three populations. Fetal, pediatric, and adult samples are represented by blue, yellow, and green bars, respectively. Inserts depict relative expression of the CYP3A family (a), CYP2C family (b), and the FMO family (c). CYP3A, cytochrome P450 3A; FMO, flavin-containing monooxygenase.
Expression of CYP3A, CYP2C, and FMO proteins in fetal, pediatric, and adult hepatic tissues. Protein expression was measured in triplicate in fetal (n=7), pediatric (n=16), and adult (n=10) HLM by LC-MRM-based targeted quantitative proteomic analysis. Panels depict protein expression of the CYP3A family (a), CYP2C family (b), and the FMO family (c). For the CYP3A family, the green, yellow, and blue squares represent CYP3A4, CYP3A5, and CYP3A7 proteins, respectively. For the CYP2C family, the aqua and purple squares represent the CYP2C9 and CYP2C19 proteins, respectively. For the FMO family, the red, orange, and grey squares depict FMO1, FMO3, and FMO5 proteins, respectively. A comparison of CYP3A (d), CYP2C (e), and FMO (f) protein expression in the fetal (open bars), pediatric (grey hatched bars), and adult (solid bars) hepatic tissues is depicted as mean values±SD. CYP3A, cytochrome P450 3A; FMO, flavin-containing monooxygenase; HLM, human liver microsome.
Expression of both CYP2C9 and CYP2C19 genes was low to undetectable in fetal samples, with approximately a 100-fold lower expression compared with that in the pediatric and adult samples (Figure 1b). No significant differences were observed in CYP2C9 or CYP2C19 gene expression between adults and children. However, there was a trend toward higher CYP2C19 gene expression in the pediatric population compared with the adult population (P=0.062). In contrast, protein expression of CYP2C9 protein increased linearly over time (r2=0.31; P<0.0001) and adult-level expression was reached in the pediatric population (Figure 2b). Overall, expression of CYP2C9 protein corresponded to its gene expression (Figure 2e); fetal tissue expressed significantly lower levels (0.001 nmol/mg) of CYP2C9 protein compared with pediatric and adult tissues (0.052 and 0.054 nmol/mg, respectively; P<0.001).
In contrast to the linear maturation profile of CYP2C9 protein, expression of CYP2C19 protein peaked during the pediatric period (Figure 2b). CYP2C19 protein expression in pediatric tissues was over twofold higher compared with adult tissues (0.0093 vs. 0.0043 nmol/mg; P=0.046). This confirms our previous observation of higher CYP2C19 protein expression in the hepatic tissue of children (2–10 years old) than in the adult hepatic tissue; however, as the previous results were based on western blot analysis, and were semiquantitative at best (21), the present results provide a more definitive estimate of the differences in the hepatic CYP2C29 protein expression between children and adult.
FMO gene expression exhibited high variability, which limited the detection of significant differences between populations or isoforms, but the trends corresponded well to those seen with protein expression. As expected, relatively high protein expression of FMO1 was observed in fetal samples (0.007 nmol/mg) with a low expression of FMO3 (0.001 nmol/mg; Figure 2c). FMO1 expression was virtually undetectable in pediatric and adult samples (0.00002 nmol/mg in each population), confirming the notion that FMO1 is a fetal isoform (11). Because of the specificity of the protein expression assay and low variability in the protein expression data, a significant difference in the expression of FMO3 protein was observed among all three populations, with an increase from 0.001 to 0.02 to 0.037 nmol/mg in the fetal, pediatric, and adult samples, respectively (P<0.05; Figure 2f). Thus, there was a clear ontogeny profile for FMO3 protein expression with a linear increase from the fetal period into adulthood (r2=0.52; P<0.0001). As seen in the CYP3A family, a distinct transition occurred from a predominant fetal isoform (FMO1) to an adult isoform (FMO3) after birth.
CYP3A, CYP2C, and FMO Enzyme Activities Normalized to Respective Protein Expression
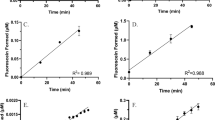
The protein expression levels of the CYP and FMO enzymes were correlated to corresponding enzyme activity using enzyme-specific probe substrates. Testosterone was chosen as the probe substrate to assess functional activity of the CYP3A family in the fetal, pediatric, and adult populations, and in recombinant CYP3A4, 3A5, and 3A7 enzymes (Figure 3). Metabolite formation was normalized to the total amount of CYP3A protein in each sample as measured by the targeted quantitative proteomic method, and a clear maturation trend was observed for the catalytic activity for oxidation of testosterone across the three populations (1.2, 1.6, and 2.8 pmol/min/pmol 3A, respectively). This maturation profile was consistent with the formation of metabolite by the recombinant 3A4, 3A5, and 3A7 enzymes (3.9, 3.0, and 1.4 pmol/min/pmol 3A, respectively).
CYP3A enzyme activity in the microsomes prepared from fetal, pediatric, and adult human hepatic tissues and in recombinant CYP3A supersomes. Fetal, pediatric, and adult HLM (n=6) and recombinant CYP3A supersomes were incubated in triplicate with testosterone and the rate of formation of 6β-hydroxytestosterone was measured. The metabolite formation was normalized to the CYP3A expressed in each sample. The horizontal line in each group represents the mean value. CYP3A, cytochrome P450 3A; HLM, human liver microsome.
Diclofenac and S-mephenytoin were used as probe substrates to assess how changes in protein expression with age affect the functional activities of CYP2C9 and CYP2C19, respectively. Formation of 4′-hydroxylation of diclofenac by fetal tissue, normalized to total microsomal protein (Figure 4a), was over 300-fold lower than that observed for the pediatric and adult samples (0.24 vs. 96 vs. 83 pmol/min/mg protein, respectively). After normalizing metabolite formation to the total CYP2C9 protein in each sample (Figure 4b), fetal samples still had much lower metabolic activity compared with pediatric and adult samples (0.2 vs. 1.6 and 1.4 pmol/mg/pmol CYP2C9, respectively), with no significant difference in CYP2C9 activity between adult and pediatric populations.
CYP2C enzyme activity in the microsomes prepared from fetal, pediatric, and adult human hepatic tissues. Fetal, pediatric, and adult HLMs (n=6) were incubated in triplicate with diclofenac to measure CYP2C9 (a,b) and with S-mephenytoin to measure CYP2C19 (c,d) activity. Metabolite formation rates were normalized to the total microsomal protein (a,c), the CYP2C9 protein (b), and the CYP2C19 protein (d). Horizontal lines represent the mean values in each panel. HLM, human liver microsome.
CYP2C19 activity displayed a different maturation profile compared with CYP2C9. Consistent with nearly undetectable CYP2C19 protein levels in fetal tissue, formation of 4′-hydroxylation of S-mephenytoin by fetal samples was undetectable (Figure 4c). However, metabolic activity in pediatric samples was higher than that in adult samples when normalized to total microsomal protein (1.6 vs. 0.51 pmol/min/mg protein, respectively; P<0.04; Figure 4c). Even after normalizing for the higher expression of CYP2C19 protein in pediatric samples, a significant difference between the metabolite formation rates by the pediatric and adult enzyme (0.15 vs. 0.079 pmol/min/pmol CYP2C19, respectively; P<0.01) was observed. Even after excluding the samples with the highest formation rates (which may be attributed to rapid metabolizers, although this is not confirmed by genetic analysis), the difference between pediatric and adult samples retained statistical significance (P<0.04). These results suggest that a yet unidentified cellular mechanism, in combination with higher CYP2C19 expression, confers higher metabolic activity in the pediatric population. Overall, CYP2C9 and CYP2C19 catalytic activities toward their respective probe substrates are significantly correlated with the abundance of each enzyme across the age groups (Figure 5).
Correlation between CYP2C protein expression and enzyme activity. Formation of 4′-OH diclofenac is correlated with the expression of CYP2C9 protein (a). Formation of 4′-OH S-mephenytoin is correlated with the expression of CYP2C19 protein (b). The graphs depict the linear correlation (solid line) and the 95th confidence intervals (dotted lines).
Functional activity of FMO was assessed using cimetidine as a probe substrate. Formation of cimetidine sulfoxide by recombinant FMO1 was twofold higher than by recombinant FMO3. Fetal samples, which had a higher expression of FMO1, generated cimetidine sulfoxide at a rate twofold higher than that observed with pediatric and adult samples (P<0.003; Figure 6a). Normalization of FMO functional activity to its abundance in the corresponding tissue revealed a surprising finding that catalytic activity of FMO is significantly higher (P<0.0001) in fetal samples (0.20 pmol/min/pmol FMO) compared with pediatric and adult samples (0.005 and 0.01 pmol/min/pmol FMO, respectively; Figure 6b).
Enzyme activity in the microsomes prepared from fetal, pediatric, and adult human hepatic tissues. Fetal, pediatric, and adult HLM (n=6) and recombinant FMO supersomes were incubated in triplicate with cimetidine to measure FMO activity. Metabolite formation rates were normalized to total microsomal protein (a) and the total FMO1 and FMO3 proteins (b). The bars represent values derived from six samples, and the error bars depict SD (a). Horizontal lines represent the mean values in each panel (b). FMO, flavin-containing monooxygenase; HLM, human liver microsome.
Discussion
It is recognized that gene and protein expression of DMEs changes with age. Although numerous studies have investigated these changes, the protein measurements have been carried out by western blot analysis, which is considered to be a semiquantitative method. In the present study, we have determined protein expression quantitatively using an LC-MS/MS method, and have related gene and protein expression to the catalytic activity of CYP3A, CYP2C, and FMO enzymes, normalized to the respective enzyme protein, in the same respective samples of the fetal, pediatric, and adult hepatic tissues. We acknowledge that the sample size is small and that range of fetal age considered was not exhaustive, and therefore the conclusions reached in this study should be verified in future studies with additional tissue samples in these age groups and expanded fetal age groups. However, our approach of relating quantitative protein expression measurements and enzyme activity in the same tissues provides reliable and self-consistent data on the metabolic changes due to the three enzyme families in the fetal, pediatric, and adult populations.
A comparison between gene and protein expression profiles indicates that gene expression can reveal relative protein expression trends among CYP isoforms within the same enzyme family. Despite this loose correlation, gene expression cannot be used as a surrogate for respective protein expression. For instance, adult CYP2C9 gene expression suggests a similar abundance to fetal CYP3A7, and a 2.5-fold higher expression compared with the adult CYP3A4 gene. However, CYP3A protein quantification reveals higher expression compared with CYP2C9, which is in agreement with published reports (23). Differences in primer specificity used to quantify gene expression can alter results, and post-translational modifications can influence protein expression, thus leading to inaccurate predictions in PK parameters for pediatric drugs if gene expression is used as a measure of protein expression.
In regards to protein expression, traditional methods (e.g., western blotting or differential metabolism) utilize antibodies that pose the likelihood of confounding the data because of cross-reactivity with other isoforms in the same family or measurement of functionally inactive apoproteins (19). However, quantitative proteomic analysis provides enhanced accuracy of enzyme abundance as target peptides are selected from an isoform’s specific peptide sequence (15). Therefore, the results presented here provide a more quantitative assessment of protein expression compared with previous studies that utilized western blot or differential metabolism to determine isoform expression within the same family (10, 13, 24).
The quantitative determination of protein expression in fetal, pediatric, and adult hepatic tissues provided the opportunity to correlate protein expression with functional activities of the DMEs in these age groups. Previous studies and reviews have reported that CYP3A7 has a lower catalytic activity (as much as 17-fold) for testosterone 6-hydroxylation compared with CYP3A4 (refs 12, 13, 22). In contrast to previously reported results, we have observed that CYP3A7 testosterone metabolism is greater than that of CYP3A4 (ref. 25). This was missed in the previous studies because the activities were compared based on 6β-OH testosterone formation, which is not the major metabolite of CYP3A7 (ref. 25). The results show that the specific activity (i.e., catalytic activity normalized to the enzyme protein) of CYP3A recombinant enzymes measured in supersomes is higher than the corresponding specific activity measured in HLM. This could be due to differences in NADPH coupling to the CYP enzyme or different lipid environments, yielding different substrate access to the enzyme.
Interestingly, protein expression of CYP2C19 was higher (nearly twofold) in pediatric liver samples compared with that in adult liver samples. This distinctive trend was observed in previous reports where CYP2C19-specific activity was ~20% higher in children aged 5 months to 10 years compared with children aged 10–18 years of age (10). Similar results regarding CYP2C19 activity differences were reported by Yanni et al. who also compared CYP2C19 protein expression using western blot analysis. They demonstrated a trend for higher expression in children (2–12 years old) compared with adults (>18 years old), but did not reach significance because of high variability (21). Although neither of these reports could definitively establish higher pediatric CYP2C19 expression, the sensitivity of the LC-MS-based targeted quantitative proteomic assay to determine CYP2C19 expression decreased variability and provided definitive evidence of a significant difference in CYP2C19 protein expression between these two populations. Higher expression of CYP2C19 in children may explain, in part, the higher pediatric clearance of voriconazole compared with adults, and previous findings that CYP2C19 accounts for higher voriconazole metabolism in children vs. adults (21, 26).
Surprisingly, when the catalytic activity of CYP2C19 was normalized to CYP2C19 protein expression, the turnover rate of the CYP2C19 probe S-mephenytoin was higher for pediatric HLM compared with adult HLM, suggesting higher catalytic efficiency of CYP2C19 in children. This significant difference remained even after excluding the samples with the highest formation rate, which could be due to CYP2C19 polymorphisms. One likely explanation for this may be a better coupling of NADPH cytochrome P450 oxidoreductase or cytochrome b5 to CYP2C19 either because of a better oxidoreductase/cytochrome b5 to CYP2C19 stoichiometry or because of different lipid environment. Structure, function, and genetic polymorphisms of NADPH cytochrome P450 oxidoreductase have been shown to affect drug metabolism of compounds such as steroids and midazolam (27, 28, 29, 30). Importantly, specific variants of NADPH cytochrome P450 oxidoreductase increased the metabolic activity of both CYP2C19 and CYP1A2 (ref. 31). As cytochrome b5 can augment cytochrome P450 activity by an alternative or additional electron transfer process or by allosteric activation (32), it is conceivable that higher specific activity of CYP2C19 in children may be caused by cytochrome b5. Another potential factor affecting catalytic efficiency of CYP2C19 is the lipid composition of the membrane within the endoplasmic reticulum. Accessibility of a cytochrome P450-active site requires a substrate to traverse the lipid membrane in which the enzyme is embedded; therefore, the same cytochrome P450 enzyme could catalyze the same substrate with different efficiencies in two different microsomal samples if lipid composition differs. As it is known that fat composition changes during development, it is conceivable that the composition of cell membranes in children may be different from that in adults. Irrespective of the mechanism, higher catalytic efficiency of CYP2C19 in the pediatric liver, in addition to higher CYP2C19 protein expression, could be responsible for the higher contribution of CYP2C19 toward voriconazole metabolism and its higher clearance in children compared with adults, which could not be accounted for by twofold greater liver-to-body mass ratio in children compared with adults. The higher catalytic efficiency of CYP2C19 in pediatric compared with adult HLM is reported for the first time in this study. Hence, we suggest that age-related differences in the molecular and biophysical determinants of the catalytic efficiency of DMEs should be further investigated. Future research should derive PK parameters of drugs using PBPK models that assume age-related changes in the catalytic efficiency of the CYP enzymes responsible for their metabolism and compared with the parameters derived using models that fix the catalytic efficiency of the CYP enzymes across different age groups.
Finally, functional FMO activity of fetal, pediatric, and adult HLM was assessed using cimetidine as a probe. Cimetidine has been studied in preclinical species as well as in human clinical studies, and the evidence thus far suggests that FMO3 is the major metabolizing enzyme of cimetidine in adult humans (33, 34). Interestingly, the results in this study show that the rate of cimetidine sulfoxide formation by fetal HLM and recombinant FMO1, normalized to FMO1 protein, was approximately twofold higher than by pediatric and adult HLM and recombinant FMO3, normalized to FMO3 protein. As was seen with CYP3A, the switch from FMO1 to FMO3 is linked to parturition rather than to gestational age (11, 35). These maturational differences in activity could also be attributed to differences in microenvironment or a coupling of the FMO proteins to their cofactor, FAD (19), and are potentially regulated by hormonal changes that occur during gestation and at birth.
Other studies have also investigated the age-dependent relationship between gene expression, protein expression, and enzyme activity. In one study by Sadler et al., which covered a larger gestational age than is considered in the present study, the authors note that it is crucial to determine pediatric activity as it is not always correlated to expression (36). The present study reaches a similar conclusion that, with the growing importance of physiologically based modeling in pediatric clinical pharmacology, it is essential to determine the activity of each enzyme in a specific age bracket in order to accurately simulate pediatric PK.
One limitation of our study is the small sample size because of limited availability of fetal and pediatric tissues, which is not unexpected in pediatric studies. On the other hand, selective and specific analytical methods were utilized to reduce variability in the measurement of protein expression. In addition, only samples with acceptable quality mRNA were included in gene expression analysis and quality, and intraday variability was assessed for protein expression. We observe that protein expression data for all three families that are reported in this study are consistent with those reported using semiquatitative methods in previous literature reports. We acknowledge that the small sample size limits the detection of differences in the expression and/or function of DMEs because of polymorphisms. As clinically significant polymorphisms have been documented for CYP2C19, CYP2C9, FMO, and CYP3A5 (refs 7, 24, 37, 38, 39), further research is warranted to understand the interaction of polymorphisms, ontogeny, expression, and activity.
Overall, the CYP3A and CYP2C families account for 60% of hepatic enzymes and metabolize over 70% of drugs currently on the market (1, 23). Although the FMO family exhibits lower expression, it has a pivotal role in regulating reactive oxygen species and metabolizing nitrogen- and sulfur-containing compounds (35). The unique differences in enzyme contributions that exist among the CYP3A, CYP2C, and FMO families during development affect pediatric PK. Although age-dependent changes in hepatic expression and activity have been examined previously, this is the first study to measure these changes based on targeted quantitative proteomics using the same samples. In addition, this is the first study, to our knowledge, that compares against one another and assesses three common techniques utilized to understand how differences in ontogeny can affect dosing in children and provides concrete evidence for the optimal method to utilize in future research. Most importantly, this research lays the groundwork for future investigations into the age-related biochemical and molecular differences as determinants of DME, which can be simulated using PBPK modeling.
References
Wienkers LC, Heath TG . Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov 2005;4:825–33.
Ginsberg G, Hattis D, Sonawane B et al. Evaluation of child/adult pharmacokinetic differences from a database derived from the therapeutic drug literature. Toxicol Sci 2002;66:185–200.
Andersson T, Hassall E, Lundborg P et al. Pharmacokinetics of orally administered omeprazole in children. International Pediatric Omeprazole Pharmacokinetic Group. Am J Gastroenterol 2000;95:3101–6.
Kanamori M, Takahashi H, Echizen H . Developmental changes in the liver weight- and body weight-normalized clearance of theophylline, phenytoin and cyclosporine in children. Int J. Clin Pharmacol Ther 2002;40:485–92.
Litalien C, Theoret Y, Faure C . Pharmacokinetics of proton pump inhibitors in children. Clin Pharmacokinet 2005;44:441–66.
Tran A, Rey E, Pons G et al. Pharmacokinetic-pharmacodynamic study of oral lansoprazole in children. Clin Pharmacol Ther 2002;71:359–67.
Leveque D, Nivoix Y, Jehl F et al. Clinical pharmacokinetics of voriconazole. Int J Antimicrob Agents 2006;27:274–84.
Hyland R, Jones BC, Smith DA . Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab Dispos 2003;31:540–7.
Yanni SB, Annaert PP, Augustijns P et al. Role of flavin-containing monooxygenase in oxidative metabolism of voriconazole by human liver microsomes. Drug Metab Dispos 2008;36:1119–25.
Koukouritaki SB, Manro JR, Marsh SA et al. Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 2004;308:965–74.
Koukouritaki SB, Simpson P, Yeung CK et al. Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr Res 2002;51:236–43.
Lacroix D, Sonnier M, Moncion A et al. Expression of CYP3A in the human liver—evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 1997;247:625–34.
Stevens JC, Hines RN, Gu C et al. Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 2003;307:573–82.
Michaels S, Wang MZ . The revised human liver cytochrome P450 "Pie": absolute protein quantification of CYP4F and CYP3A enzymes using targeted quantitative proteomics. Drug Metab Dispos 2014;42:1241–51.
Ohtsuki S, Uchida Y, Kubo Y et al. Quantitative targeted absolute proteomics-based ADME research as a new path to drug discovery and development: methodology, advantages, strategy, and prospects. J Pharm Sci 2011;100:3547–59.
Wang MZ, Wu JQ, Dennison JB et al. A gel-free MS-based quantitative proteomic approach accurately measures cytochrome P450 protein concentrations in human liver microsomes. Proteomics 2008;8:4186–96.
Fisher CD, Lickteig AJ, Augustine LM et al. Hepatic cytochrome P450 enzyme alterations in humans with progressive stages of nonalcoholic fatty liver disease. Drug Metab Dispos 2009;37:2087–94.
Kawakami H, Ohtsuki S, Kamiie J et al. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J Pharm Sci 2011;100:341–52.
Chen Y, Zane NR, Thakker DR et al. Quantification of flavin-containing monooxygenases 1, 3 and 5 in human liver microsomes by UPLC-MRM-based targeted quantitative proteomics. Drug Metab Dispos 2015;44:975–83.
Daniele G, Fieu M, Joachim S et al. Rapid analysis of diclofenac and some of its transformation products in the three-spined stickleback, Gasterosteus aculeatus, by liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem 2016;408:4435–4444.
Yanni SB, Annaert PP, Augustijns P et al. In vitro hepatic metabolism explains higher clearance of voriconazole in children vs adults: role of CYP2C19 and flavin-containing monooxygenase 3. Drug Metab Dispos 2010;38:25–31.
Ince I, Knibbe CA, Danhof M et al. Developmental changes in the expression and function of cytochrome P450 3A isoforms: evidence from in vitro and in vivo investigations. Clin Pharmacokinet 2013;52:333–45.
Shimada T, Yamazaki H, Mimura M et al. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 1994;270:414–23.
Koukouritaki SB, Poch MT, Cabacungan ET et al. Discovery of novel flavin-containing monooxygenase 3 (FMO3) single nucleotide polymorphisms and functional analysis of upstream haplotype variants. Mol Pharmacol 2005;68:383–92.
Zane NR Predicting Pharmacokinetic Behavior and Dose of Sildenafil and Voriconazole in Neonatal and Pediatric Populations by In Vitro Metabolism and PBPK Modeling. Thesis, The University of North Carolina, 2015.
Zane NR, Thakker DR . A physiologically based pharmacokinetic model for voriconazole disposition predicts intestinal first-pass metabolism in children. Clin Pharmacokinet 2014;53:1171–82.
Chen X, Pan LQ, Naranmandura H et al. Influence of various polymorphic variants of cytochrome P450 oxidoreductase (POR) on drug metabolic activity of CYP3A4 and CYP2B6. PLoS ONE 2012;7:e38495.
Oneda B, Crettol S, Jaquenoud Sirot E et al. The P450 oxidoreductase genotype is associated with CYP3A activity in vivo as measured by the midazolam phenotyping test. Pharmacogenet Genomics 2009;19:877–83.
Pandey AV, Fluck CE . NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol Ther 2013;138:229–54.
Pandey AV, Sproll P . Pharmacogenomics of human P450 oxidoreductase. Front Pharmacol 2014;5:103.
Agrawal V, Huang N, Miller WL . Pharmacogenetics of P450 oxidoreductase: effect of sequence variants on activities of CYP1A2 and CYP2C19. Pharmacogenet Genomics 2008;18:569–76.
Porter TD . The roles of cytochrome b5 in cytochrome P450 reactions. J Biochem Mol Toxicol 2002;16:311–6.
Cashman JR, Park SB, Berkman CE et al. Role of hepatic flavin-containing monooxygenase 3 in drug and chemical metabolism in adult humans. Chem Biol Interact 1995;96:33–46.
Cashman JR, Park SB, Yang ZC et al. Chemical, enzymatic, and human enantioselective S-oxygenation of cimetidine. Drug Metab Dispos 1993;21:587–97.
Krueger SK, Williams DE . Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 2005;106:357–87.
Sadler NC, Nandhikonda P, Webb-Robertson BJ et al. Hepatic cytochrome P450 activity, abundance, and expression throughout human development. Drug Metab Dispos 2016;44:984–91.
Weiss J, Ten Hoevel MM, Burhenne J et al. CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol 2009;49:196–204.
Shirasaka Y, Chang SY, Grubb MF et al. Effect of CYP3A5 expression on the inhibition of CYP3A-catalyzed drug metabolism: impact on modeling CYP3A-mediated drug-drug interactions. Drug Metab Dispos 2013;41:1566–74.
Jacobson PA, Oetting WS, Brearley AM et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 2011;91:300–8.
Acknowledgements
Author contributions
Participated in research design: N.R.Z., Y.C., M.Z.W., and D.R.T. Conducted experiments and performed analysis: N.R.Z. and Y.C. Wrote or contributed to the writing of the manuscript: N.R.Z., Y.C., M.Z.W., and D.R.T.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Statement of Financial Support
This work was supported in part by the United States National Institutes of Health (R01GM089994), and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (Award number 5 T32 GM086330-04).
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary material is linked to the online version of the paper at
Supplementary information
Rights and permissions
About this article
Cite this article
Zane, N., Chen, Y., Wang, M. et al. Cytochrome P450 and flavin-containing monooxygenase families: age-dependent differences in expression and functional activity. Pediatr Res 83, 527–535 (2018). https://doi.org/10.1038/pr.2017.226
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2017.226
This article is cited by
-
Overexpression of the salicylic acid binding protein 2 (SABP2) from tobacco enhances tolerance against Huanglongbing in transgenic citrus
Plant Cell Reports (2022)
-
Accidental intoxications in toddlers: lack of cross-reactivity of vilazodone and its urinary metabolite M17 with drug of abuse screening immunoassays
BMC Clinical Pathology (2019)
-
A repository of protein abundance data of drug metabolizing enzymes and transporters for applications in physiologically based pharmacokinetic (PBPK) modelling and simulation
Scientific Reports (2019)
-
Voriconazole: A Review of Population Pharmacokinetic Analyses
Clinical Pharmacokinetics (2019)