Abstract
Background:
The aim was to identify susceptibility alleles for infantile hypertrophic pyloric stenosis (IHPS) in a pedigree previously linked to IHPS5 on chromosome 16q24.
Methods:
We screened the positional and functional candidate gene FOXF1 by Sanger sequencing in a single affected individual. All family members for whom DNA was available were genotyped to determine cosegregation status of the putative causal variant. Immunofluorescence studies were performed to compare the cellular localization of wildtype and mutant form of the protein. Transcriptional activity was compared using a luciferase assay.
Results:
A single novel substitution in FOXF1 (c.416G>A) predicted to result in a missense mutation (R139Q) was shown to cosegregate with disease trait. It was not seen in 560 control chromosomes nor has it been reported in ExAC or ESP. The R139Q substitution affects a conserved arginine residue within the DNA-binding domain of FOXF1. The transcriptional activity of the mutant FOXF1 protein is significantly reduced in comparison to wild-type.
Conclusion:
These results provide strong evidence that the R139Q substitution in FOXF1 causes IHPS in this family and imply a novel pathological pathway for the condition. They further support a role for FOXF1 in the regulation of embryonic and neonatal development of the gastro-intestinal tract.
Similar content being viewed by others
Main
Infantile hypertrophic pyloric stenosis (IHPS, MIM # 179010) is the most common inherited cause of gastrointestinal obstruction in the first few months of life with an incidence of 1–8 per 1,000 live births in the Caucasian population. It typically affects infants 3–12 wk after birth (1) with clinical features including projectile vomiting, weight loss, and dehydration. Treatment is by pyloromyotomy, which was introduced a century ago (see review by MacMahon 2006 (2)).
A genetic predisposition is clearly established, although environmental factors are also important (2). A prone sleeping position has been proposed and investigated as a potential cause of IHPS. This theory stemmed from the observation of parallel decreasing incidences of IHPS and sudden infant death syndrome (SIDS) (3). However, the most recent longitudinal study does not support this theory (4). There is population-based evidence supporting bottle-feeding as a risk factor (5). Pre- and postnatal exposure to erythromycin have been proposed as risk factors for IHPS but studies are not consistent although the risk associated with postnatal exposure appears to be more significant (6,7,8,9).
IHPS shows familial aggregation and represents a paradigm for the multifactorial sex-modified threshold model of inheritance, with affected males outnumbering females in a 4:1 ratio (10). IHPS is predicted to be oligogenic, determined by two or three loci of moderate effect estimated to confer individual genotype relative risks of up to 5 (11).
IHPS has been associated with several genetic syndromes (12,13) and chromosomal abnormalities (14,15). Autosomal dominant monogenic forms of IHPS have also been reported in several extended pedigrees (16,17,18). Five loci for familial IHPS have been identified: IHPS1 (the NOS1 gene on chromosome 12q24) (19); IHPS2 (chromosome 16p12-p13) (16); IHPS3 (chromosome 11q14-q22) (20); IHPS4 (chromosome Xq23) (20); and IHPS5 (chromosome 16q24) (21).
IHPS5 (OMIM #612525) was identified through a SNP-based whole genome linkage scan of a single extended family with multiple cases of IHPS; the family also described in this paper. Chromosome 16 was the only chromosome for which a significant logarithm of odds (LOD) score (>3) was achieved, assuming autosomal dominant inheritance with reduced penetrance. A shared haplotype on one chromosome extends across 4.2Mb of the linked region and is present in all affected individuals and obligate carriers on whom genotyping was performed (seven cases and two obligate carriers; Supplementary Table S1 online). This region of chromosome 16 contains a cluster of FOX genes: FOXF1; FOXC2; and FOXL1. FOX proteins are transcription factors characterized by the classic winged-helix-turn-helix (wHTH) ~110 amino acid binding domain. In most cases the protein is composed of three α-helices (H1-H3), two to four β sheets and two loops (the “wings”, W1 and W2) (see review by Carlsson and Mahlapuu, 2002 (22). FOX proteins tend to bind as monomers with H3 acting as the recognition helix to fill the major groove of the target DNA while W2 interacts with the minor groove (23,24). Previous work has shown that mutations affecting the amino acid sequence of the W2 domain can affect transactivation ability (25).
In humans, mutations in FOXF1 have been associated with a range of GI tract abnormalities including duodenal stenosis, annular pancreas, congenital short bowel, and intestinal malrotation (26). It has been shown in mice that during organogenesis, Foxf1 is expressed in splanchnic mesoderm which gives rise to the muscular components of the GI tract, such as the pyloric smooth muscle (27,28). There is evidence that Foxf1 is a downstream effector of the Sonic Hedgehog pathway with respect to lung, stomach, and intestinal development. These data support FOXF1 as an excellent positional and functional candidate gene for IHPS.
The aim of our work was to identify susceptibility alleles in the linked region on chromosome 16. Here, we report the identification of a missense variant in FOXF1 which segregates with disease in the large extended IHPS family previously mapped to 16q24.3. We demonstrate that the variant reduces the transcriptional efficacy of the resulting protein in vitro. We suggest that this variant may cause IHPS in this family by affecting downstream expression of other genes.
Methods
Subjects and Samples
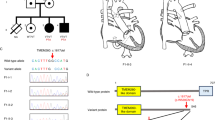
A large Irish Caucasian family was ascertained as previously described (16); all affected individuals had undergone pyloromyotomy for IHPS (see Figure 1 for pedigree structure). The study was approved by the University College London Hospital Ethics Committee and relevant local regional committees. Informed consent was obtained from all participating individuals.
Schematic showing the pedigree structure of family IHPS78 and the SNP and microsatellite genotypes across the linked region. Squares indicate males; circles females; a line through indicates that the individual is deceased. Shaded individuals are affected with infantile hypertrophic pyloric stenosis (IHPS). The disease-associated haplotype is indicated by the brown bar. Other haplotypes are of varying style simply to differentiate between them; there is no significance to the color or style. Those individuals who are heterozygous for the c.416G>A mutation are indicated with a black asterisk.
Fine-Mapping of Associated Haplotype
Three microsatellite markers were selected to reduce the size of the associated haplotype. These markers were: D16S520, D16S3074 and D16S3077. Oligonucleotide primers were designed to amplify these microsatellites in all members of the family on whom DNA was available. Primer details were taken directly from UniSTS (primer and protocol details available on request). Supplementary Table S1 online contains a list of the SNPs and microsatellites which comprise the associated haplotype and their relative positions.
Candidate Gene Analysis
FOXF1. The FOXF1 gene has two coding exons totaling 1140 nucleotides. Oligonucleotide primers were designed to amplify these two exons and the exon–intron boundaries in three fragments (primer details available on request). A single affected individual from family 78 (III.9) was resequenced to identify putative causal variants. Mutation Taster (29) and Polyphen2 (30) were used to predict the biological effect of any putative causal variant identified. The ClustalW alignment tool was used to compare protein sequences (31).
The remaining family members from whom DNA was available were genotyped for the identified putative causal variant using a restriction digest utilizing the restriction enzyme MspA1I and/or direct Sanger sequencing of the surrounding area.
This variant was screened for in the resource of affected IHPS patients (549 individuals, both sporadic and familial cases) and in 280 controls by outsourcing to KBiosciences (now part of LGC Genomics).
Approximately 5 kb of genomic DNA, extending from 500 bp upstream of the start of the 5’UTR of FOXF1 to 500 bp downstream of the end of the 3’UTR of FOXF1, was resequenced in 58 affected individuals from the largest 58 IHPS pedigrees in our resource. This resequencing was outsourced to Polymorphic DNA Technologies, Inc.
FOXC2 and FOXL1. FOXC2 has a single coding exon totaling 1,683 nucleotides and FOXL1 has a single coding exon totaling 3,189 nucleotides. These exons plus 500 bp upstream and downstream were resequenced in a single affected individual from the 39 families consistent with linkage to 16q24.3 (as identified through the previous genome-wide linkage studies (20,21); again primers are available on request). This resequencing was performed in-house. A putative causal variant in FOXL1 was genotyped in remaining family members using the restriction enzyme DrdI.
Expression Constructs
A FOXF1 expression construct was the kind gift of Prof. P. Carlsson (32). The FOXF1-p.R139Q (FOXF1-mut) mutation was synthesized by Eurofins MWG Operon and verified by sequencing. The FOXF1 and FOXF1-mut constructs were sub-cloned into both pCDNA3 and pCMV-Myc (Clontech). Details of mutagenesis and sub-cloning are available on request. A FOXF1 luciferase reporter construct, pGHV(-158/+57)-luc, containing upstream sequences from Growth Hormone Variant was the kind gift of Dr. S. Handwerger (33).
Luciferase Assays
HepG2 cells (ATCC Number HB 8065) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal calf serum, penicillin, and streptomycin and grown at 37 °C in a humidified atmosphere of 5% CO2. Cells were plated at 25,000 cells per well so that they were between 40–70% confluent 24 h later. Transfections were performed in sextuplicate using Polyfect (QIAGEN, Manchester, UK) according to manufacturer’s instructions. Each well contained 50 ng of the reporter construct, 500 ng of pCDNA3 (containing FOXF1, FOXF1-mut or β-galactosidase as control), and 10 ng of pRL-TK (Promega, Southampton, UK) for normalization. An empty luciferase vector (pGL3, Promega) was used as a control. Normalized luciferase activity was measured after 48 h using a Dual-Luciferase assay system (Promega). Results are a representative example from one experiment. Statistical analysis was performed using GraphPad Prism software (GraphPad Software, La Jolla, CA). Samples were compared by one-way ANOVA with Tukey’s post-comparison test. Values are expressed as means ± SEM.
Immunofluorescence
HEK-293 cells (ATCC Number CRL-1573) were cultured and transfected using the same conditions as above, except that cells were plated onto Poly-D-lysine coated glass cover slips in a 24-well plate and 2 µg of pCMV-Myc DNA was used. Cells were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde 24 h after transfection. The myc epitope tag was detected using a rabbit-anti-myc antibody (Abcam ab9106) at 1:500 dilution and a goat-anti-rabbit Alexa Fluor 488 (Invitrogen, Carlsbad, CA) secondary antibody at 1:200 dilution. Nuclei were counterstained with Hoechst 33258.
Results
Fine-Mapping of Associated Haplotype
We first attempted to fine map the IHPS5 region using three microsatellite markers. Figure 1 shows the genotypes for all single nucleotide polymorphisms (SNPs) within the associated haplotype and the three microsatellites genotyped for this report. Genotyping of the microsatellite markers did not reduce the size of the critical region. As such, all three FOX genes remained functional and positional candidates requiring further investigation.
Candidate Gene Analysis
FOXF1. Resequencing of individual III.9 identified a single novel missense variant in exon 1 of FOXF1 (NM001451.2); c.416G>A (counting the A of the start codon as position 1), p.R139Q (counting the start codon as residue number 1). The patient was heterozygous for this substitution (Supplementary Figure S1 online; see also Supplementary Table S2 online for other known SNPs identified). Analysis using Mutation Taster predicted this amino acid change to be disease causing. Polyphen also predicts this change to be “Probably Damaging” (with a score of 1.00).
The presence of the A allele was shown to create an MspA1I site (the recognition site for MspA1I is CMGCKG; in the wildtype, this sequence is CGGCGG which would not be recognized by the enzyme; the mutation changes this sequence to CAGCGG which is recognized by the enzyme and the fragment is cut). Individuals homozygous for the wild type G allele have a single fragment 531 bp in length; GA heterozygotes have two fragments, one 531 bp, the other 442 bp in size; individuals who are AA homozygotes would have just the 442 bp fragment. All affected members genotyped (III.1, III.9, III.14, IV.1, IV.2, and IV.4), either through a restriction digest or through direct Sanger sequencing, were heterozygous for the variant (GA). Both obligate carriers (III.2 and III.11) were also heterozygous for the mutation ( Figure 1 ). All unaffected, noncarriers genotyped (II.4, II.5, III.3, III.5, III.6, III.7, III.10, III.13, III.15, and IV.5) were homozygous for the wild type allele. The remaining family members could not be genotyped due to unavailability of DNA.
This variant has not been reported in either ExAC or the Exome Sequencing Project and we did not identify it through sequencing of 560 control chromosomes.
FOXC2 and FOXL1. Resequencing identified a number of SNPs but no putative causal variants (Supplementary Tables S3 and S4 online).
FOXF1 Trans-Activation Activity
The p.R139Q substitution found in individual III.9 affects a conserved arginine residue within the DNA-binding domain of FOXF1 and is predicted to impair protein function. FOXF1 has been shown previously to act as a transcriptional activator (32,33). In order to compare the trans-activation activity of FOXF1 and FOXF1-p.R139Q, we used a luciferase reporter construct, pGHV-luc, containing control elements from the 5’-flanking region of growth hormone variant (33). Transient transfection of FOXF1 constructs into HepG2 cells did not activate a promoter-less pGL3 reporter. The pGHV-luc reporter showed increased, basal activity in HepG2 cells. Co-transfection with FOXF1 caused robust activation of pGHV-luc (~5-fold), whereas the FOXF1-p.R139Q variant showed no significant effect on luciferase activity ( Figure 2 ).
Trans-activation of pGHV-luc by wild type and mutant FOXF1. Wild type FOXF1 (black fill) induces robust activation of pGHV(-158/+57)-luc (pGHV), but FOXF1-p.R139Q (gray fill) does not. Background activity is seen with an empty pCMV-myc vector (white fill). Neither FOXF1 construct affects the activity of a promoter-less pGL3 vector (pGL3). Values are expressed as relative luciferase units (RLU), normalized to pRL-TK and represent the mean ± SEM of six replicates. †P < 0.01.
FOXF1 Protein Localization
The lack of trans-activation activity of FOXF1-p.R139Q suggests that this variant represents a loss of function mutation. To exclude the possibility that the p.R139Q substitution causes mis-targeting of the FOXF1 protein, we generated N-terminal, myc epitope-tagged fusion proteins for both FOXF1 and FOXF1-p.R139Q by sub-cloning into pCMV-Myc. Constructs were transiently transfected into HEK-293 cells and protein localization detected by immunofluorescence using an anti-myc antibody. Nuclei were counter-stained with Hoechst dye. Both FOXF1 and FOXF1-p.R139Q proteins were localized to the nucleus, but not the cytoplasm ( Figure 3 ), demonstrating that the p.R139Q variant is stable and correctly targeted to the nucleus.
Subcellular localization of wild type and mutant FOXF1. Myc-epitope tagged FOXF1 proteins were detected in HEK-293 cells by immunofluorescence using a rabbit-anti-myc antibody and goat-anti-rabbit Alexa Fluor 488 secondary antibody (b,e). Nuclei were counter-stained with Hoechst 33258 (a,d). FOXF1 localization appears nuclear in the merged images (c), as expected. FOXF1-p.R139Q localization also appears nuclear in the merged images (f) and is not affected by the missense substitution.
Discussion
In this study, we have identified a novel nucleotide substitution (c.416G>A) in FOXF1 which segregates with disease and carrier status in a single family with IHPS and reduces the trans-activation ability of the protein. Though this appears to be a rare, family-specific mutation, it does provide important new scientific information on IHPS and FOXF1 function.
Family IHPS78 has a number of interesting features. The two known “obligate carriers” share with affected individuals a single copy of the disease-associated haplotype within which lies the single copy of the mutant allele c.416G>A. The presence of obligate carriers suggests that the mutation is not fully penetrant or that there may be variable expressivity. Diagnosis of IHPS can usually be made on clinical examination with a typical history of forceful vomiting. This can be confirmed by either upper gastrointestinal contrast study or ultrasound examination which is now routinely performed. IHPS can be considered as a continuous trait as measured by the hypertrophy of the pyloric musculature. Babies with IHPS can have variable degree of vomiting determined by the severity/degree of the pyloric muscle hypertrophy and obstruction. Infants with diagnostic features of IHPS on radiological examinations yet without significant symptoms have been reported. It is likely that mild cases of IHPS exist who never require medical intervention and resolve spontaneously by 3–4 mo of age. IHPS is a complex condition which arises from interaction between multiple predisposing genes and environmental factors. The obligate carriers in this family most likely represent mild cases of IHPS who may not have had all the necessary or sufficient genetic and/or environmental triggers required during the appropriate timeframe to develop clinically diagnosable IHPS. It is plausible that affected individuals share genetic variants at other loci that may interact with the FOXF1 mutation leading to disease expression while obligate carriers do not share these other interacting variants. However, it should be pointed out that this family did not show linkage to any of the other reported familial IHPS loci so any interaction is likely to be with variants at other loci. For example, a genome-wide association study has identified three variants significantly associated with IHPS which have implicated the genes MBNL1 and NKX2-5 (34). Two of these variants were shown to be associated in an independent Northern European replication cohort (35). The variant associated with NKX2-5 was also significantly associated in a Chinese cohort (36). More recently, a further locus was implicated by genome-wide association analyses that indicate a possible role for a gene on chromosome 11q23.3 (37) and there is the suggestion that variants in RET may play a contributory role in IHPS (38). Clearly IHPS has a highly complex, multifactorial mode of inheritance involves multiple predisposing alleles and different biological pathways, as well as environmental cues. Modeling these interactions is an interesting challenge that may become possible as we understand more about the etiology. Typically, there is a male to female predominance in IHPS but this is not seen in this family (there are five males affected and three females; of the obligate carriers, one is male and one is female). This is also observed in other large pedigrees with multiple affected in our resource, i.e., in families with many cases of IHPS there are more equal numbers of male and female cases. In our resource of 358 trios, the ratio of male to female cases is approximately 5:1 while among our 141 multiple case pedigrees, it is around half this at ~5:2. It is not understood why more males are affected by IHPS than females but it seems likely that males are somehow more biologically predisposed and that their risk threshold is lower (one of the linked loci, IHPS4, is on the X chromosome which may yet explain the gender bias). However, when there is a genetic variant which is sufficiently severe, the likelihood of females also passing their higher risk threshold increases and therefore families occur with multiple cases and although still more males are affected, the difference is not so apparent. In this situation, gender has less effect on disease risk. It is plausible that the biological significance of alternative genetic variants differ considerably depending upon the gene (and pathway) affected leading to differing levels of penetrance. Whether the same equality of gender distribution is seen in other families with multiple cases of IHPS may depend on which gene contains the causal variant.
Results of our study confirm an important role for FOXF1 in the development and function of upper gastrointestinal tract. The substitution results in a change in the amino acid sequence (p.R139Q). This residue is conserved in murine Foxf1 as well as many FOX proteins including those encoded by the chromosome 16 cluster (Supplementary Figure S2 online). This residue is also conserved in FOXF1 homologues across several species (data not shown except for mouse but available via HomoloGene). The exact structure of the FOXF1 protein has not been compiled but based on known structures of other related FOX proteins and on amino acid sequence alignment (22,25), residue 139 of FOXF1 is likely to form part of the W2 domain. This region of the protein is responsible for binding to the shallow groove of the target DNA. We show that c.416G>A (p.R139Q) affects the normal function of the FOXF1 protein by the reporter gene assay. The trans-activation activity of the mutant FOXF1 is statistically significantly reduced in comparison to the wildtype and is comparable to that of the empty vector. Our immunofluorescence studies show that the mutant FOXF1 is localizing normally to the nucleus. Our hypothesis therefore is that the mutant FOXF1 trafficks normally to the nucleus but binds to the target binding sequence with reduced efficacy. How this eventually leads to IHPS has not been answered by this study but will continue to be part of our future work.
It has been established that murine Foxf1 is a target gene of the Hedgehog (Hh) pathway (39,40) which is a key component of the development of the vertebrate gut. Haploinsufficiency of Foxf1 causes lung and gut malformations (40). During embryonic development, Foxf1 is primarily expressed in the splanchnic mesoderm from which the smooth musculature of the GI tract is derived (22). In humans, inactivating mutations and microdeletions of FOXF1 have been associated with alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) and a range of malformations in the heart, lung and GI tract including intestinal malrotation and duodenal stenosis (26). None of these features is present in any member of family IHPS78 and GI tract abnormalities are not seen in all patients with ACDMPV. This may be because this family only has a missense mutation in FOXF1 which does not lead to complete inactivation of the protein. However, recently a missense mutation at the same site (R139L) was identified in three siblings with ACDMPV, the segregation of which appeared to be consistent with paternal imprinting of FOXF1 (41). Our results are not consistent with this but differential patterns of imprinting have been reported between different tissues (42) which could partly explain the contrasting results with regards to severity of disease and mode of inheritance.
Two other genes in the chromosome 16q24.1 cluster of FOX genes are FOXC2 and FOXL1. We could not exclude them based on our haplotype analysis so we also resequenced these genes in IHPS patients but did not find any putative causal mutations which segregated with disease. We acknowledge that there are other genes in this region which we did not resequence and it is possible that the variant we have identified in FOXF1 is in linkage disequilibrium with the actual causal variant which may reside in another gene. However, FOXF1 is the best functional candidate in the region and the evidence from our luciferase assay supports the hypothesis that the p.R139Q mutation in FOXF1 is causal in this family.
There appear to be potentially many downstream targets of FOXF1 including growth hormone variant (GHV), as utilized within our study during the luciferase assay, but also including members of the S1P/S1PR1 signaling pathway (43) and genes involved in lung development and angiogenesis (44). In mice, it has been shown that FoxF1 acts either independently or as part of a reciprocal pathway with BMP4 to trigger expression of Gata4, which itself forms part of a family of transcription factors important in development (45). It is often thought that the smooth muscle hypertrophy seen in IHPS is a result of failure of relaxation which suggests that FOXF1 normally inhibits the action of a downstream target. The identification of the specific targets of FOXF1 affected in this instance will be a major focus of our immediate future work.
There still remain many unanswered questions regarding the age and tissue specific nature of IHPS and function of FOXF1 in relation to this which are beyond the scope of this preliminary study but will need to be investigated further.
In summary, we have identified FOXF1 as a new disease gene for IHPS and suggest a novel biological pathway to be investigated to further our understanding of the disorder. The coexistence of a mutation in FOXF1 and IHPS also provides further evidence for a role for FOXF1 in embryonic, and possibly neonatal, development of the GI tract.
Supplemental Data Description
Supplementary data contains four tables pertaining to: the SNPs and microsatellites which constitute the associated haplotype across chromosome 16q24.3; and details of the variants identified through resequencing of FOXF1, FOXC2 and FOXL1. Two figures are also within the Supplementary data: a figure showing the wildtype sequence around the variant and the variant in heterozygous state; and a figure showing conservation of the residue affected by the variant in FOXF1 across the different FOX proteins.
Accession Numbers
The identified variant has been submitted to dbSNP and has recently been given an identifier: rs672601295.
Statement of Financial Support
No extramural financial support was received in support of this work.
Disclosure
None of the authors have any conflict of interest, financial or otherwise, to declare.
References
Aboagye J, Goldstein SD, Salazar JH, et al. Age at presentation of common pediatric surgical conditions: Reexamining dogma. J Pediatr Surg 2014;49:995–9.
MacMahon B. The continuing enigma of pyloric stenosis of infancy: a review. Epidemiology 2006;17:195–201.
Persson S, Ekbom A, Granath F, Nordenskjöld A. Parallel incidences of sudden infant death syndrome and infantile hypertrophic pyloric stenosis: a common cause? Pediatrics 2001;108:E70.
de Laffolie J, Turial S, Heckmann M, Zimmer KP, Schier F. Decline in infantile hypertrophic pyloric stenosis in Germany in 2000-2008. Pediatrics 2012;129:e901–6.
McAteer JP, Ledbetter DJ, Goldin AB. Role of bottle feeding in the etiology of hypertrophic pyloric stenosis. JAMA Pediatr 2013;167:1143–9.
Lin KJ, Mitchell AA, Yau WP, Louik C, Hernández-Díaz S. Safety of macrolides during pregnancy. Am J Obstet Gynecol 2013;208:221.e1–8.
Lozada LE, Royall MJ, Nylund CM, Eberly MD. Development of pyloric stenosis after a 4-day course of oral erythromycin. Pediatr Emerg Care 2013;29:498–9.
Lund M, Pasternak B, Davidsen RB, et al. Use of macrolides in mother and child and risk of infantile hypertrophic pyloric stenosis: nationwide cohort study. BMJ 2014;348:g1908.
Sørensen HT, Skriver MV, Pedersen L, Larsen H, Ebbesen F, Schønheyder HC. Risk of infantile hypertrophic pyloric stenosis after maternal postnatal use of macrolides. Scand J Infect Dis 2003;35:104–6.
Carter CO. The inheritance of congenital pyloric stenosis. Br Med Bull 1961;17:251–4.
Mitchell LE, Risch N. The genetics of infantile hypertrophic pyloric stenosis. A reanalysis. Am J Dis Child 1993;147:1203–11.
Jackson L, Kline AD, Barr MA, Koch S. de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet 1993;47:940–6.
Smith DW, Lemli L, Opitz JM. A newly recognised syndrome of multiple congenital anomalies. J Pediatr 1964;64:210–7.
Heller A, Seidel J, Hübler A, et al. Molecular cytogenetic characterisation of partial trisomy 9q in a case with pyloric stenosis and a review. J Med Genet 2000;37:529–32.
Hodgson SV, Berry AC, Dunbar HM. Two brothers with an unbalanced 8;17 translocation and infantile pyloric stenosis. Clin Genet 1995;48:328–30.
Capon F, Reece A, Ravindrarajah R, Chung E. Linkage of monogenic infantile hypertrophic pyloric stenosis to chromosome 16p12-p13 and evidence for genetic heterogeneity. Am J Hum Genet 2006;79:378–82.
Finsen VR. Infantile hypertrophic pyloric stenosis–unusual familial incidence. Arch Dis Child 1979;54:720–1.
Fried K, Aviv S, Nisenbaum C. Probable autosomal dominant infantile pyloric stenosis in a large kindred. Clin Genet 1981;20:328–30.
Chung E, Curtis D, Chen G, et al. Genetic evidence for the neuronal nitric oxide synthase gene (NOS1) as a susceptibility locus for infantile pyloric stenosis. Am J Hum Genet 1996;58:363–70.
Everett KV, Chioza BA, Georgoula C, et al. Genome-wide high-density SNP-based linkage analysis of infantile hypertrophic pyloric stenosis identifies loci on chromosomes 11q14-q22 and Xq23. Am J Hum Genet 2008;82:756–62.
Everett KV, Capon F, Georgoula C, et al. Linkage of monogenic infantile hypertrophic pyloric stenosis to chromosome 16q24. Eur J Hum Genet 2008;16:1151–4.
Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol 2002;250:1–23.
Cirillo LA, Zaret KS. Specific interactions of the wing domains of FOXA1 transcription factor with DNA. J Mol Biol 2007;366:720–4.
Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta 2011;1813:1946–53.
Murphy TC, Saleem RA, Footz T, Ritch R, McGillivray B, Walter MA. The wing 2 region of the FOXC1 forkhead domain is necessary for normal DNA-binding and transactivation functions. Invest Ophthalmol Vis Sci 2004;45:2531–8.
Stankiewicz P, Sen P, Bhatt SS, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet 2009;84:780–91.
Mahlapuu M, Pelto-Huikko M, Aitola M, Enerbäck S, Carlsson P. FREAC-1 contains a cell-type-specific transcriptional activation domain and is expressed in epithelial-mesenchymal interfaces. Dev Biol 1998;202:183–95.
Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech Dev 1997;69:53–69.
Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010;7:575–6.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9.
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947–8.
Hellqvist M, Mahlapuu M, Samuelsson L, Enerbäck S, Carlsson P. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J Biol Chem 1996;271:4482–90.
Lomenick JP, Hubert MA, Handwerger S. Transcription factor FOXF1 regulates growth hormone variant gene expression. Am J Physiol Endocrinol Metab 2006;291:E947–51.
Feenstra B, Geller F, Krogh C, et al. Common variants near MBNL1 and NKX2-5 are associated with infantile hypertrophic pyloric stenosis. Nat Genet 2012;44:334–7.
Everett KV, Chung EM. Confirmation of two novel loci for infantile hypertrophic pyloric stenosis on chromosomes 3 and 5. J Hum Genet 2013;58:236–7.
Feng Z, Liang P, Li Q, Nie Y, Zhang Y. Association between NKX2-5 rs29784 and infantile hypertrophic pyloric stenosis in Chinese Han population. Int J Clin Exp Med 2015;8:2905–10.
Feenstra B, Geller F, Carstensen L, et al. Plasma lipids, genetic variants near APOA1, and the risk of infantile hypertrophic pyloric stenosis. JAMA 2013;310:714–21.
Serra A, Schuchardt K, Genuneit J, et al. The role of RET genomic variants in infantile hypertrophic pyloric stenosis. Eur J Pediatr Surg 2011;21:389–94.
Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J Biol Chem 2009;284:5936–44.
Mahlapuu M, Enerbäck S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development 2001;128:2397–406.
Sen P, Gerychova R, Janku P, et al. A familial case of alveolar capillary dysplasia with misalignment of pulmonary veins supports paternal imprinting of FOXF1 in human. Eur J Hum Genet 2013;21:474–7.
Baran Y, Subramaniam M, Biton A, et al.; GTEx Consortium. The landscape of genomic imprinting across diverse adult human tissues. Genome Res 2015;25:927–36.
Cai Y, Bolte C, Le T, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal 2016;9:ra40.
Sen P, Dharmadhikari AV, Majewski T, et al. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PLoS One 2014;9:e94390.
Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development 2005;132:3405–17.
Acknowledgements
We would like to thank all the families who agreed to take part in the study without whom none of this work would be possible. We would also like to thank Prof Mark Gardiner for all his advice and encouragement regarding this project and many others; as well as for his proof-reading of the manuscript. None of the authors have any conflict of interest, financial or otherwise, to declare.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures and tables
(DOCX 3390 kb)
Rights and permissions
About this article
Cite this article
Everett, K., Ataliotis, P., Chioza, B. et al. A novel missense mutation in the transcription factor FOXF1 cosegregating with infantile hypertrophic pyloric stenosis in the extended pedigree linked to IHPS5 on chromosome 16q24. Pediatr Res 81, 632–638 (2017). https://doi.org/10.1038/pr.2016.244
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2016.244
This article is cited by
-
Co-occurrence of infantile hypertrophic pyloric stenosis and congenital heart defects: a nationwide cohort study
Pediatric Research (2019)
-
Pyloric stenosis: an enigma more than a century after the first successful treatment
Pediatric Surgery International (2018)