Abstract
Two polymorphisms in the murine double minute 2 (MDM2) gene (rs1690916 and rs2279744) have been associated with the risk of osteosarcoma (OS). When we analyzed these two polymorphisms in two new independents cohorts (Spanish and Slovenian), we found no association. In order to clarify this, we conducted a meta-analysis including six populations, with a total of 246 OS patients and 1,760 controls for rs1690916; and 433 OS patients and 1,959 controls for rs2279744. Pooled odds ratio risks and corresponding 95% CI were estimated to assess the possible associations. Our results showed that these two polymorphisms were not associated with the susceptibility of OS under any genetic model studied. In conclusion, the present meta-analysis indicates that MDM2 rs1690916 and rs2279744 cannot be considered as genetic risk factors for OS susceptibility in the different populations. Therefore, the influence of these two polymorphisms on the risk of OS may be less important than previously suggested. Future studies are needed to confirm these results.
Similar content being viewed by others
Main
Osteosarcoma (OS) is the most common primary malignant tumor of bone, mainly occurring in the second decade of life. The precise etiology of the disease remains partially unknown (1). Nevertheless, genetic factors might play a key role in its pathogenesis (2). To date diverse studies have reported associations of common genetic variants in biologically plausible pathways with OS risk (3,4,5,6). Among the analyzed variants, rs1690916 and rs2279744 in the murine double minute 2 gene (MDM2) are two of the most recurrently studied and associated with the susceptibility of OS (5,7,8,9). The MDM2 gene is especially interesting because it functions as an E3 ubiquitin ligase promoting p53 degradation (10,11).
MDM2 rs1690916, located at the 3′ untranslated region of the gene, was significantly associated with the risk of OS in a large North-American population including 96 patients and 1,416 controls (7). They found that rs1690916 AA genotype decreased the risk of the disease. rs1690916 A allele was also found to be associated with a decreased risk of bone tumors in Russian population including 68 patients and 86 controls (9). However, this study only included 26 OS patients out of 68 analyzed. MDM2 rs2279744, situated in the promoter region, was related to an increased risk of OS (GG vs. TT) in Italian population (211 OS patients and 250 controls) (5), being this association stronger in females. Moreover, a robust correlation between rs2279744 G allele enrichment and MDM2 amplification was found, suggesting the contribution of this polymorphism to OS tumorigenesis (12). However, this single nucleotide polymorphism (SNP) did not show any association in the North-American population (7). A meta-analysis evaluating the association between these two polymorphisms and the risk of OS was performed, concluding that both SNPs influence the risk of OS (13). Nevertheless, some inaccuracies were detected in the study. Among others, the lack of information in the methods used to select the studies and extract the genotyping data and the inclusion of other types of bone tumors in the meta-analysis (not only OS) (14,15).
Therefore, this study aimed to clarify the possible association between MDM2 rs1690916 and rs2279744, and risk of OS. We initially analyzed the effect of both polymorphisms in two new independent cohorts of OS patients (Spanish and Slovenian) and performed a new updated meta-analysis following the recommended guidelines published (16).
Results
Association Study in Spanish and Slovenian Populations
The genotyping success rate was 96.5% for rs1690916 (126 patients and 258 controls) and 94.5% for rs2279744 (120 patients and 256 controls). Genotype frequencies in controls were consistent with those expected from Hardy–Weinberg equilibrium (P > 0.05).
The allelic and genotypic association analyses showed that none of the two MDM2 polymorphisms were associated with OS susceptibility neither in the Spanish, nor in the Slovenian population (P > 0.05, under all the genetic models used) ( Table 1 and Supplementary Table S1 online). When both set of samples were analyzed together, no significant association with OS susceptibility was observed in either set (data not shown).
Meta-Analysis
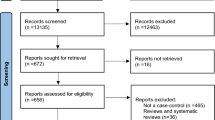
The original search provided 720 records. After eliminating duplications, 693 records remained. Of these, 670 were discarded after reviewing the abstracts because they did not meet the required criteria for inclusion. The full texts of the remaining 23 studies were examined in detail. Of these, we identified a total of four studies that investigated the association between MDM2 SNPs rs1690916 (n = 2) and rs2279744 (n = 3) and the risk of OS ( Figure 1 ).
Flow-chart of study selection.
The populations analyzed were Russian (9), American (7), Australian (12) and Italian (5). We added Spanish and Slovenian datasets to the meta-analysis. The characteristics of the six studies are presented in Table 2 . The distribution of genotypes in the controls of each study was in agreement with Hardy–Weinberg equilibrium (P > 0.05).
MDM2 rs1690916 polymorphism. The meta-analysis on rs1690916 included a total of four populations with 246 OS patients and 1,760 controls. Among them, the American population showed a significant association with OS risk, while the other three populations did not show association. Overall, the meta-analysis results indicated that this polymorphism was not statistically associated with OS risk under any genetic model studied (P > 0.05) ( Figure 2 ). The heterogeneity of studies on this polymorphism was > 75%, under allele, codominant, and dominant models. The individual study’s influence on the pooled results was also analyzed showing that no study affected the pooled OR significantly (see Supplementary Table S2 online).
Forest plot for meta-analysis of the association between rs1690916 polymorphism and osteosarcoma risk under (a) allele model (b) codominant model (c) recessive model and (d) dominant model.
MDM2 rs2279744 polymorphism. A total of five populations including 433 OS patients and 1,959 controls were analyzed in the meta-analysis on rs2279744. Four showed no significant results while one (Italian) showed an increased risk of OS under all genetic models (allele, codominant, recessive, and dominant). The meta-analysis results displayed that rs2279744 polymorphism was not associated with the susceptibility to OS (P > 0.05) ( Figure 3 ).
Forest plot for meta-analysis of the association between rs2279744 polymorphism and osteosarcoma risk under (a) allele model (b) codominant model (c) recessive model and (d) dominant model.
Publication bias. The shapes of funnel plot did not reveal obvious evidence of asymmetry ( Figure 4 ), and all the P-values of Egger’s tests were >0.05, providing statistical evidence of the funnel plots’ symmetry (see Supplementary Figures S1 and S2 online). This indicates that biases from publication may not have influence on the results.
Funnel plots of the Egger’s test of allele comparison for publication bias. (a) rs1690916 A vs. G comparison (b) rs2279744 G vs. T comparison.
Discussion
Association between the MDM2 polymorphisms rs1690916 and rs2279744 and OS risk has been studied but still needs to be clarified. In this study, we analyzed the effect of these two polymorphisms in two new independent cohorts of OS patients, and no significant results were detected. We also performed a meta-analysis with all available data including six populations with a total of 464 OS and 2,048 controls. The results showed that rs1690916 and rs2279744 did not increase the risk of OS.
MDM2 polymorphisms rs1690916 and rs2279744 have been widely studied in relation to the susceptibility of OS because of their putative functional effect: rs16909616 is located at 3′ untranslated region of MDM2 and rs2279744 (also known as SNP309) is situated in the promoter. rs2279744 has been shown to increase the affinity of the transcription factor Sp1, incrementing the MDM2 basal level (17), which in turn could decrease the p53 response leading to the development of the tumor.11 Our association study, however, showed no relationship between these two polymorphisms and risk of OS in two independent cohorts (Spanish and Slovenian). Moreover, we observed in the meta-analysis that OR values in our two populations were in opposite direction from the previous studies, suggesting the existence of population differences leading to different SNP effects. These differences could be explained by gene environmental interactions.
In line with our results, a recent genome wide association study in OS did not find association with MDM2 (3). Since these negative results were different from previous published results, we decided to perform a deep revision of data published in the literature and realized a new meta-analysis. The results showed no association, in contrast to another meta-analysis previously published, (13) in which some inaccuracies were detected (14,15). The differences between both meta-analyses could be due to the fact that our study included two new populations, but they can also be explained because different data from the original articles were selected. In the case of rs1690916, we specifically extracted from the Russian population (9) the genotype data of the OS patients upon request, observing no significant association. In the case of rs2279744 in Australian population (12), we used as controls benign tumors. After doing the association analyses we found no significant association.
Taken all these data in consideration, our meta-analysis has several strengths. First, we used a comprehensive search strategy with well-defined inclusion and exclusion criteria. Second, two reviewers performed the study selection and data extraction independently and discrepancies were resolved by consensus. Third, we assessed the quality of the included studies by predefined criteria and the score of included studies here was high (scored at least 5, in 5 out of 6 studies). Finally, all genotype data extracted from the studies are reported in the meta-analysis. Nevertheless, there are still some limitations. First, although we included two new populations to the meta-analysis, the number of cases is still relatively small; which is inherent to the low incidence of this tumor. Second, the heterogeneity of the studies was very high in the case of rs1690916. We explored the sources of this heterogeneity analyzing if any of the studies individually affected the pooled OR. However, none of them altered significantly the pooled OR, indicating that other unknown factors might be the cause. Finally, although we are reluctant to include patients with benign tumors as controls in susceptibility studies, we decided to include the Australian population mainly because it had been included in the previous meta-analysis (13). In any case, we checked the results with and without this population and they did not vary significantly (data not shown).
In conclusion, the present meta-analysis indicates that MDM2 rs1690916 and rs2279744 are not genetic risk factors for OS susceptibility in the different populations. Therefore, the influence of these two polymorphisms on the risk of OS may be less important than previously suggested. Future studies are needed to confirm these results.
Methods
Association Study
Study population. Two new independent cohorts of OS patients from Spain (n = 99) and Slovenia (n = 40) and their corresponding controls (n = 167 and n = 92; respectively) were analyzed. The Spanish patients came from the Hospital University La Paz Madrid, the Hospital University Donostia and the Department of Genetics, Physical Anthropology and Animal Physiology of UPV/EHU. The Slovenian patients were diagnosed at the Department of Hematology and Oncology, University Children’s Hospital, Ljubljana, Slovenia, or at the Institute of Oncology, Ljubljana, Slovenia (18). All cases were diagnosed by experienced pathologists and oncologists. Unrelated healthy donors were used as control groups in both populations. Informed consent was obtained from all patients or their parents before sample collection. The study was approved by the local ethics committees (105/2009 and 67/02/12) and was carried out according to the Declaration of Helsinki.
Genotyping. Genomic DNA was extracted from peripheral blood using standard procedures. Peripheral blood samples were obtained as the source of DNA from Spanish patients and all healthy controls, while in Slovenian OS patients DNA was extracted from the areas of formalin fixed, paraffin embedded material verified by an experienced pathologist to be representative of normal tissue. The genotyping analyses were performed by using amplification-refractory mutation system polymerase chain reaction (ARMS)-polymerase chain reaction for rs1690916 and polymerase chain reaction followed by restriction analysis with MspA1I enzyme for rs2279744. Duplicates were included in each assay. The polymerase chain reaction products were visualized after electrophoresis on 3% agarose gels. Primer sequences and polymerase chain reaction conditions are described in detail in Supplementary Table S3 online.
Systematic Review and Meta-analysis
Search strategy. We performed an exhaustive search to identify studies that examined the association between the rs1690916 and rs2279744 polymorphisms of MDM2 and OS susceptibility. We used the keywords and subject terms (“bone tumor” OR osteosarcoma) AND (polymorphism* OR SNP*), (“bone tumor” OR osteosarcoma) AND (MDM2 OR “murine double minute 2”), (rs1690916 OR rs2279744 OR T309G) for PubMed database (National Center for Biotechnology Information, US National Library of Medicine, Bethesda, MD) searches for articles published until December 2015. All references cited in the studies were then reviewed to possibly identify additional publications.
Inclusion and exclusion criteria. Original studies that investigated the association between rs1690916 and rs2279744 polymorphisms and OS risk with sufficient data to calculate crude OR values were included. Reviews, meta-analyses and studies analyzing other regions or variants were excluded.
Data extraction. For each article, we gathered year of publication, first author, country of origin, ethnicity of population, sample size and genotype and/or allele frequencies. All data were independently extracted by two investigators and reached conformity on all items through consultation. When it was not possible to extract the genotype data from the article, we contacted the authors to obtain them.
Quality assessment. The quality of included studies was assessed independently by two investigators by scoring according to a “methodological quality assessment scale” (see Supplementary Table S4 online), which was modified from previous meta-analyses (19,20). In the scale, five items, including the representativeness of cases, source of controls, sample size, quality control of genotyping methods and Hardy–Weinberg equilibrium were carefully checked. Quality scores ranged from 0 to 10 and a higher score indicated better quality of the study. Scores > 5 were considered acceptable.
Statistical Analysis
The data were statistically processed by R version 2.15 software R Foundation for Statistical Computing, Vienna, Austria (http://www.R-project.org). Genotype frequencies in cases and controls were compared using a χ2-test. The deviation from Hardy–Weinberg equilibrium was also calculated by a χ2-test (in the healthy population). The effect sizes of the associations were estimated by the odds ratios (OR) from univariate logistic regression using different genetic models. In all cases the significance level was set at 5%. For the meta-analysis, we used an additive model. The overall pooled OR and corresponding 95%CI were estimated using Mantel–Haenszel’s method, with random effects model. The heterogeneity was quantified using the I2 statistic (0–25% no heterogeneity, 25–50% moderate heterogeneity, 50–75% large heterogeneity, and 75–100% extreme heterogeneity). To evaluate the publication bias of literatures in this meta-analysis, Begg’s funnel plot and Egger’s test were performed (21). These tests analyze the intervention effect estimates from individual studies against some measure of each study’s size or precision. This means that effect estimates from small studies will therefore scatter more widely at the bottom of the graph, with the spread narrowing among larger studies. In absence of bias, the plot should resemble a symmetrical funnel. If there is bias, the plot will have an asymmetrical appearance.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Statement of Financial Support
This study was funded by the Basque Government (IT661-13), UPV/EHU (UFI11/35) and RTICC (RD12/0036/0060, RD12/0036/0036). All the authors declare no conflict of interest.
References
Rosenberg AE, Cleton-Jansen AM, de Pinieux G, Deyrup AT, Hauben E, Squire J. Conventional osteosarcoma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, eds. WHO Classification of Tumours of Soft Tissue and Bone. 4th edn. Lyon, France: IARC Press, 2013: 282–8.
Hameed M, Dorfman H. Primary malignant bone tumors–recent developments. Semin Diagn Pathol 2011;28:86–101.
Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet 2013;45:799–803.
Ruza E, Sotillo E, Sierrasesúmaga L, Azcona C, Patiño-García A. Analysis of polymorphisms of the vitamin D receptor, estrogen receptor, and collagen Ialpha1 genes and their relationship with height in children with bone cancer. J Pediatr Hematol Oncol 2003;25:780–6.
Toffoli G, Biason P, Russo A, et al. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin Cancer Res 2009;15:3550–6.
Savage SA, Burdett L, Troisi R, Douglass C, Hoover RN, Chanock SJ ; National Osteosarcoma Etiology Study Group. Germ-line genetic variation of TP53 in osteosarcoma. Pediatr Blood Cancer 2007;49:28–33.
Mirabello L, Yu K, Berndt SI, et al.; National Osteosarcoma Etiology Study Group. A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer 2011;11:209.
He J, Wang J, Wang D, et al. Association analysis between genetic variants of MDM2 gene and osteosarcoma susceptibility in Chinese. Endocr J 2013;60:1215–20.
Naumov VA, Generozov EV, Solovyov YN, Aliev MD, Kushlinsky NE. Association of FGFR3 and MDM2 gene nucleotide polymorphisms with bone tumors. Bull Exp Biol Med 2012;153:869–73.
Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997;387:296–9.
Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol 2011;12:439–52.
Ito M, Barys L, O’Reilly T, et al. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res 2011;17:416–26.
Wang L, Liu Z, Jing P, et al. Effects of murine double minute 2 polymorphisms on the risk and survival of osteosarcoma: a systemic review and meta-analysis. Tumour Biol 2014;35:1649–52.
Bilbao-Aldaiturriaga N, Martin-Guerrero I, Garcia-Orad A. Letter regarding Wang et al. entitled “Effects of murine double minute 2 polymorphisms on the risk and survival of osteosarcoma: a systematic review and meta-analysis”. Tumour Biol 2014;35:6179.
Liu G, Xu W, Hao Y, Xu Z. Letter regarding Wang et al. entitled “Effects of murine double minute 2 polymorphisms on the risk and survival of osteosarcoma: a systemic review and meta-analysis”. Tumour Biol 2014;35:3943–4.
The Cochrane Collaboration. Higgins JPT, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0, 2011. (http://handbook.cochrane.org/)
Knappskog S, Lønning PE. MDM2 promoter SNP285 and SNP309; phylogeny and impact on cancer risk. Oncotarget 2011;2:251–8.
Goričar K, Kovač V, Jazbec J, Lamovec J, Dolžan V. Homologous recombination repair polymorphisms and the risk for osteosarcoma. J Med Biochem 2014; 33:1–7.
Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One 2012;7:e30585.
Li K, Tie H, Hu N, et al. Association of two polymorphisms rs2910164 in miRNA-146a and rs3746444 in miRNA-499 with rheumatoid arthritis: a meta-analysis. Hum Immunol 2014;75:602–8.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34.
Acknowledgements
The Slovenian Osteosarcoma Study Group members contributing patients data to this study: Katja Goričar (Institute of Biochemistry, Faculty of Medicine, Ljubljana), Viljem Kovač (Pharmacogenetics Laboratory, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana), Janez Jazbec (Institute of Oncology Ljubljana), Janez Lamovec (Oncology and Hematology Unit, University Children’s Hospital, University Medical Centre, Ljubljana).
Author information
Authors and Affiliations
Consortia
Corresponding author
Supplementary information
Supplementary Tables
(DOC 98 kb)
Supplementary Figure S1
(TIFF 34 kb)
Supplementary Figure S2
(TIFF 32 kb)
Rights and permissions
About this article
Cite this article
Bilbao-Aldaiturriaga, N., Askaiturrieta, Z., Granado-Tajada, I. et al. A systematic review and meta-analysis of MDM2 polymorphisms in osteosarcoma susceptibility. Pediatr Res 80, 472–479 (2016). https://doi.org/10.1038/pr.2016.120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2016.120