Abstract
Mechanisms underlying the increased risk of recurrent wheeze after respiratory syncytial virus lower respiratory tract infection (RSV LRTI) are unclear. Specifically, information about genetic determinants of recurrent wheeze after RSV LRTI is limited. We performed a candidate gene association study to identify genetic determinants of recurrent wheeze after RSV LRTI. We investigated 346 single nucleotide polymorphisms (SNPs) in 220 candidate genes in 166 Dutch infants hospitalized for RSV LRTI. Logistic regression analysis was used to study associations between genotypes and haplotypes and recurrent wheeze after RSV LRTI. We found associations with recurrent wheeze for SNPs in IL19, IL20, MUC5AC, TNFRSF1B, C3, CTLA4, CXCL9, IL4R, and IL7 genes. Haplotype analysis of the combined IL19/IL20 genotyped polymorphisms demonstrated an inverse association between the TGG haplotype and recurrent wheeze after RSV LRTI. IL19 and IL20 genes were notably associated with recurrent wheeze in infants without asthmatic parents. The association of IL20 SNP rs2981573 with recurrent wheeze was confirmed in a healthy birth cohort. We concluded that genetic variation in adaptive immunity genes and particularly in IL19/IL20 genes associates with the development of recurrent wheeze after RSV LRTI, suggesting a role for these IL10 family members in the etiology of airway disease during infancy.
Similar content being viewed by others
Main
Respiratory syncytial virus lower respiratory tract infection (RSV LRTI) during infancy is an independent risk factor for subsequent recurrent wheeze, at least within the first years of childhood (1). Mechanisms underlying the increased incidence of wheeze during the first years after RSV LRTI are unclear. Recurrent wheeze after RSV LRTI was related to signs of airflow limitation during RSV LRTI (2), eosinophilia during RSV LRTI (3), and monocyte IL10-production during the convalescent phase of RSV LRTI (4).
To date, only two studies aimed to identify genetic determinants of recurrent wheeze after RSV LRTI. In one association study of 134 RSV hospitalized infants, a variant of the IL8 gene was related to the development of subsequent wheeze (5). We previously demonstrated an association between a functional IL13 polymorphism and wheeze at age 6, whereas no association was found between the IL13 polymorphism and recurrent wheeze during the first year after RSV LRTI (6).
The availability of analytic tools to study larger numbers of genes and a larger cohort of RSV LRTI-hospitalized infants in whom recurrent wheeze was evaluated enabled us to extend our previous studies. Herein, we describe the results of 346 genotyped single nucleotide polymorphisms (SNPs) on 210 genes, including IL10 family genes.
METHODS
Subjects and design.
The infants included in this study participated in previous studies (4,6–8). In brief, they were hospitalized for RSV LRTI during the winter seasons of 1995–1996 or 2004–2006. Infants included in the winter season of 1995–1996 participated in an observational study investigating the development of recurrent wheeze after RSV LRTI (4,6,7) and infants included in the 2004–2006 seasons received placebo medication in a placebo-controlled trial investigating the role of inhaled beclomethasone to prevent the occurrence of recurrent wheeze after RSV LRTI (8). Identical inclusion criteria were used in both original studies. Infants were hospitalized on suspicion of RSV LRTI, and RSV infection was confirmed by a positive RSV immunofluorescence in nasopharyngeal cells. We included previously healthy infants, i.e. infants with a history of cardiac or pulmonary disease were excluded. For this study, we selected infants of native Dutch origin who participated in the follow-up programs and whose parents prospectively recorded the presence of wheeze in a daily log (2). Identical logs were used in both original studies. The primary outcome of this study was predefined as the presence of wheeze during the first 15 mo after RSV LRTI hospitalization (8). We chose this duration of follow-up to capture the second winter season, which showed high incidence of wheeze after RSV LRTI in a previous study (9). Infants who wheezed more than the median counted days with wheeze during the follow-up of 15 mo, i.e. 14 or more d, were (arbitrarily) classified as infants with recurrent wheeze; whereas infants who wheezed less than the median counted days with wheeze during follow-up, i.e. less than 14 d, were classified as infants without recurrent wheeze. All parents provided written, informed consent. The Ethics Review Committee of the University Medical Centre Utrecht and other participating centers approved the study.
DNA isolation, genotyping, and selection of SNPs.
A candidate gene approach was followed as described in our previous study focusing on genetic determinants of severe acute RSV LRTI (7). Briefly, 384 SNPs in 220 genes were selected based on literature studies in the context of RSV infection and classified into five processes, i.e. the airway mucosal response, innate immunity, chemotaxis, adaptive immunity, and allergic asthma. DNA isolation and genotyping of patients and their parents was performed in our previous study (7). SNPs were genotyped using Illumina's Beadarray technology on a 384 Sentrix array matrix. All SNPs were assessed to determine whether the observed genotype frequencies reflected the measured allele frequencies with Hardy-Weinberg equilibrium using χ2 tests (p < 0.01) in control subjects not hospitalized because of RSV LRTI (7). All SNPs were examined for their minor allele frequency (MAF >10%) and call rate (call rate ≥90%). Thirty-eight SNPs were excluded because of low signal, overlapping of multiple clusters, or scattering of the clusters.
Replication cohort.
To confirm our main finding, we genotyped IL20 SNP rs2981573 (c.379–152 A→G), IL19 SNP rs2243188 (c.552 + 49 C→A), and IL19 SNP rs2243191 (Ser213Pro) in 90 infants recruited from an ongoing prospective unselected birth cohort of healthy newborns (10,11). The replication cohort was unselected with regard to RSV LRTI. Infants born to women delivering vaginally at term after uncomplicated pregnancy and delivery were recruited. Recurrent wheeze was measured during the first year of life using identical prospective daily recordings as used in the RSV cohort. Other genetic polymorphisms were not tested in this cohort.
Statistics.
We used logistic regression analysis to estimate the OR for genotypes associated with recurrent wheeze after RSV LRTI (SPSS for Windows, Release 15.0: SPSS Inc., Chicago, IL). Significance was set at p < 0.05. If less than five infants in either the “recurrent wheeze” or “no recurrent wheeze” groups were homozygous for the minor allele, these infants were analyzed together with heterozygous infants. X-linked SNPs were analyzed separately in boys and girls. We performed sensitivity analyses for observed significant associations in which infants with and without recurrent wheeze were distinguished according to alternative cutoff values [e.g. no wheeze at all (n = 29) versus any wheeze during follow-up (n = 137); the quartile of infants with most frequent recurrent wheeze, i.e. more than 49 d during follow-up (n = 42) versus the rest (n = 124)]. Because baseline differences existed between infants with and without recurrent wheeze, we performed post hoc stratified analyses for groups of infants with and without asthmatic parents (i.e. parental reported physician diagnosed asthma) and for groups of infants with and without signs of airflow limitation during acute RSV LRTI (i.e. physician diagnosed wheezing by auscultation).
The global test for groups of genes was used to determine whether the groups of genes involved in different immunological processes, as preclassified in our previous study, were associated with recurrent wheeze after RSV LRTI (7,12). Haplotype analysis was performed in regions with moderate to high-linkage disequilibrium (LD) (0.3–0.8) where multiple SNPs were associated with recurrent wheeze. Pairwise LD was estimated using Haploview (version 4.0, released 21 August 2007, http://www.broad.mit.edu/mpg/haploview), and extent of LD was expressed in terms of standardized R2 characteristics. Parental and infant SNP information was used to estimate haplotypes of the infants (Unphased software, version 3.0.7) (13). Haplotypes occurring with a frequency of ≥5% were included in haplotype analyses. Logistic regression analysis was used to estimate the OR for haplotypes associated with recurrent wheeze after RSV LRTI. The false discovery rate (FDR) method by Benjamini and Hochberg (14), accepting 5% false discoveries, was used to correct for testing multiple hypotheses.
RESULTS
One hundred sixty-six infants were included in this study. The median of counted days with wheeze during follow-up was 14 d (range, 0–279 d). The pattern of wheeze after RSV LRTI in the 1995–1996 and the 2004–2006 cohorts was remarkably similar. Baseline characteristics of infants with and without recurrent wheeze are presented in Table 1. Infants with recurrent wheeze more frequently exhibited signs of airflow limitation during RSV LRTI (63.2 versus 44.2%, p = 0.02), and more parents of infants with recurrent wheeze tended to suffer from asthma (16.9 versus 7.2%, p = 0.06).
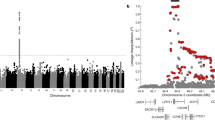
Genotype determination was successful for 346 SNPs. Ten SNPs in nine genes were associated with recurrent wheeze at the genotype level (p < 0.05). Results of the genotype-phenotype association study are presented in Table 2. The global test for groups of genes was used to evaluate the importance of the selected processes in susceptibility to recurrent wheeze after RSV LRTI. The group of SNPs in genes involved in adaptive immunity was associated with recurrent wheeze (p = 0.03) whereas the other processes were not. Six SNPs within the group of SNPs in genes involved in the adaptive immune system were significantly associated with recurrent wheeze after RSV LRTI. The three associated SNPs in the IL19 and IL20 genes were in moderate to high LD with each other (Fig. 1). To test whether individual protective effects of IL19 and IL20 polymorphisms could be attributed to a specific haplotypic background, haplotype analysis of the IL19 and IL20 genes was executed. A combined haplotype analysis was performed with two of the three genotyped SNPs in the IL19 and IL20 genes that were associated with recurrent wheeze [IL19 SNP rs2243191 (Ser213Pro) and IL20 SNP rs2981573 (c.379–152 A→G)] and with one IL20 SNP that was not associated with recurrent wheeze after RSV LRTI [IL20 SNP rs2981572 (c.-1053 T→G)]. The IL19 SNP rs2243188 (c.552 + 49 C→A) was excluded because of high LD (R2 = 0.89) with IL19 SNP rs2243191 (Ser213Pro). Three common haplotypes with a frequency ≥5% were identified in the total group of infants (Table 3). These haplotypes comprised 99% of all IL19/IL20 haplotypes. The combined IL19/IL20 haplotype TGG had a lower frequency in infants with recurrent wheeze compared with infants without recurrent wheeze [13 versus 29%; OR, 0.4 (95% CI, 0.2–0.8); p = 0.003].
The genes of the IL10 family on chromosome 1. IL10, IL19, IL20, and IL24 genes on chromosome 1 and the genotyped SNPs. SNPs rs2243188, rs2243191, and rs2981573 showed significant association with recurrent wheeze after RSV LRTI. Pairwise LD between SNPs is characterized in terms of standardized R2 characteristics. Black blocks indicate high LD between SNPs; dark and light gray blocks indicate moderate to low LD; and white blocks indicate that there is little significant LD.
Baseline differences in the presence of signs of airflow limitation during RSV LRTI and the presence of parental asthma existed between infants with and without recurrent wheeze (Table 1). To test whether associations between the IL19 and IL20 SNPs and recurrent wheeze differed for infants with and without an atopic predisposition and for infants with and without signs of airflow limitation, post hoc stratified analyses were performed. Post hoc stratification for the presence of signs of airflow limitation did not alter the associations (data not shown). Post hoc stratification for the presence of parental asthma showed that associations between IL19 and IL20 SNPs and recurrent wheeze were limited to the major subgroup of infants without asthmatic parents (Table 4). No association was observed between the IL19 and IL20 SNPs and recurrent wheeze in the minor subgroup of infants with asthmatic parents. Similar effect sizes were obtained when the analyses were stratified for other atopic features in parents, i.e. infants with and without parents suffering from hay fever and eczema (data not shown).
Sensitivity analyses in which infants with and without recurrent wheeze were distinguished according to alternative cutoff values revealed comparable results (data not shown). To determine whether the associations between the three IL19/IL20 SNPs and recurrent wheeze were limited to children with a history of RSV LRTI, we studied the three SNPs in a small unselected prospective birth cohort using identical log-based methodologies as used in the RSV cohort to quantify infant wheeze during the first year of life (10,11). The IL20 SNP rs2981573, IL19 SNP rs2243188, and IL19 SNP rs2243191 were genotyped in 90 infants. IL20 SNP rs2981573 was significantly associated with recurrent wheeze during the first year of life (OR, 0.39; 95% CI, 0.16–0.96, p = 0.04). For others SNPs, we could not confirm an association with recurrent wheeze, although similar trends were observed for both IL19 SNPs rs2243191 (OR, 0.64; 95% CI, 0.26–1.53, p = 0.31) and rs2243188 (OR, 0.74; 95% CI, 0.31–1.74, p = 0.49) and for the IL19/IL20 TGG haplotype (OR, 0.69; 95% CI, 0.37–1.27, p = 0.15). Post hoc stratification for the presence of parental asthma showed similar results (data not shown).
DISCUSSION
This study demonstrates that genetic variation in adaptive immunity genes and particularly in IL19 and IL20 genes seems to be associated with the occurrence of recurrent wheeze after RSV LRTI. The prevalence of recurrent wheeze was lower in infants with the combined IL19/IL20 TGG haplotype compared with infants with the CTA haplotype. The relationship between the IL20 SNP rs2981573 and recurrent wheeze was confirmed in a small healthy birth cohort.
We previously demonstrated the importance of SNPs in innate immune genes to determine susceptibility to RSV LRTI (7). These genes are not associated with the development of recurrent wheeze after RSV LRTI, which we now show to be determined by variation in IL10-related genes. Relationship between the IL10 family member genes IL19 and IL20 and recurrent wheeze after RSV LRTI or any other chronic airway disease have not yet been described in literature. Of the two other studies that reported on genetic susceptibility of recurrent wheeze after RSV LRTI, one study showed no association (6). The other study of Goetghebuer et al. (5) reported an association between the IL8 −251 C→T polymorphism and recurrent wheeze, which we could not confirm. However, Goetghebuer et al. analyzed the occurrence of wheeze after RSV LRTI in infants with a mean age of 6.5 y, whereas our study focused on recurrent wheeze during the first year after RSV LRTI only. These differences in wheeze phenotypes might have influenced the results because our previous study suggested that recurrent wheeze during the first year after RSV LRTI and recurrent wheeze at the age of 6 y are distinct entities with distinct immunological and genetic characteristics (6).
The major strength of our study is that polymorphisms in genes involved in different biological pathways were studied in a cohort of RSV LRTI hospitalized infants that was prospectively followed to evaluate the occurrence of recurrent wheeze. Some of our findings deserve further discussion.
First, the presence of false-positive results cannot be precluded because most of the observed associations lost significance after correction for multiple testing using the FDR method by Benjamini and Hochberg (14). However, based on the number of associated SNPs in a process, genes involved in adaptive immunity were overrepresented. Furthermore, the association of IL19 SNP rs2243191 and IL20 SNP rs2981573 with recurrent wheeze in the major subgroup of infants without asthmatic parents remained significant after FDR correction. A haplotype analysis of the SNPs on the IL19/IL20 region showed association of the TGG haplotype and recurrent wheeze, potentially pointing at a functional variant located on this haplotype. Finally, we confirmed and expanded our conclusion on the association between the IL20 SNP rs2981573 and recurrent wheeze in a replication cohort that was unselected for RSV LRTI.
Second, the post hoc observation that associations between IL19 and IL20 SNPs and recurrent wheeze were particularly detected in infants without an atopic background gave the impression that IL19 and/or IL20 cytokines are involved in nonatopic wheeze during infancy. The baseline observation that parental asthma was more common in infants with recurrent wheeze might refer to the heritability of atopic wheeze. It is known that RSV LRTI is a risk factor for subsequent recurrent wheeze independent of atopic status (1). We hypothesize that IL19 and IL20 cytokines are predominantly involved in nonatopic viral-induced recurrent wheeze. This hypothesis is further supported by a recent trial demonstrating reduced wheeze after antibody-mediated RSV prevention in nonatopic but not in atopic preterm infants (15). However, in our study, interaction terms between IL19 and IL20 SNPs and atopic features did not reach significance (Table 4). In addition, this study is relatively small and weak genetic effects may remain undetected. The OR of the observed genetic associations with recurrent wheeze was 0.4 (IL19 SNP rs2243191 and IL20 SNP rs2981573) and 0.5 (IL19 SNP rs2243188), respectively. Using QUANTO 1.1 (16), we calculated that the power to detect associations with these effect sizes was 75% (OR, 0.4) and 54% (OR, 0.5), respectively, in this study. The power to detect smaller genetic effects in children with or without asthmatic parents is low, and therefore lack of significant association does not preclude a smaller, still relevant, association.
Third, this study aimed to explain recurrent wheeze after RSV LRTI but does not address the relationship between LRTI caused by other viruses and development of reactive airway disease. It is of particular interest that the IL20 SNP rs2981573 association with recurrent wheeze was replicated in a cohort that was unselected for RSV LRTI. This might signify that IL19 and IL20 genes have a role in infant wheeze in the general population, potentially regardless of RSV LRTI. For instance, rhinovirus-associated wheezing illness is strongly linked to recurrent wheeze and allergic asthma development (17). New studies are required to study the role of genetic variation in IL19 and IL20 genes and recurrent wheeze after LRTI caused by other viruses including rhinovirus.
IL19 and IL20 are members of the IL10 family that were initially identified during a sequence database search aimed to find potential IL10 gene homologs (18,19). IL10 is a pleiotropic anti-inflammatory cytokine known to suppress Th1-like immune responses and promote Th2 responses (20). IL10 levels measured during acute RSV LRTI related to disease severity in one study (4), whereas another study showed no association (21). We previously showed that monocyte IL10 production during the convalescent phase of RSV LRTI is predictive of the subsequent development of recurrent wheeze (4). Monocyte IL10 levels measured during acute RSV LRTI were not associated with the IL19/IL20 haplotypes in a subgroup of 40 patients (data not shown). Several studies focused on the role of genetic variation in the IL10 gene locus in the pathophysiology of acute RSV LRTI. Overall, the frequency of IL10 polymorphisms in infants with RSV LRTI did not differ from controls (22–25). However, in infants hospitalized ≤6 mo of age, the IL10 −592C allele was related to RSV LRTI hospitalization (23). In addition, genetic variation at the IL10 gene locus was associated with the need for mechanical ventilation (24) and with the frequency of pneumonia (25) in RSV LRTI-hospitalized infants. The SNPs that were associated with recurrent wheeze after RSV LRTI in this study, i.e. particularly SNPs in adaptive immunity genes, were not associated with acute RSV LRTI (7), suggesting that RSV LRTI and the subsequent occurrence of recurrent wheeze have a different genetic etiology.
IL19 and IL20 genes are clustered together with IL10 and IL24 genes on chromosome 1q31–32 and have similar genomic structures and similar primary and secondary protein structures (26). Both IL19 and IL20 bind to the IL20 receptor complex, consisting of the IL20R1 and IL20R2 subunits. IL20 also binds to a heterodimeric receptor consisting of IL22R1 and IL20R2 (27). The receptors for IL19 and IL20 are widely expressed, but only lung and skin tissue express both receptors (28). Both receptors signal through STAT3 (18,27). IL10 family members cross-regulate expression of other IL10 family members. IL19 induces selective expression of IL10 by monocytes and myeloid dendritic cells (29). IL19 induces IL19 expression by an auto-feedback mechanism, which is not yet fully understood. Control of IL19 expression is provided by IL10, strongly interfering with IL19 gene transcription. The IL19 and IL20 genes contain a highly polymorphic, informative repeat sequence useful for genotyping (30). Genotyped SNPs in this study are located in the intron, exon, and promoter region. Only the IL19 SNP rs2243191 resulted in an amino acid change, i.e. Ser → Pro. Previous studies showed associations of the genotyped IL19 and IL20 SNPs with Hepatitic C virus clearance (31), psoriasis (32), palmoplantar pustulosis (33), and juvenile idiopathic arthritis (34), suggesting that IL19 and IL20 play a role in the pathology of inflammatory disorders. It is still to be determined whether the polymorphisms have differential effects on the function of the encoded protein or levels of gene expression and thus contribute to disease etiology. Limited data were available on the role of IL19 and IL20 in the etiology of airway diseases. In asthmatics, IL19 serum levels are increased, but no human data on levels in bronchoalveolar lavages have been published (35). In mice and humans, IL19 overexpression enhanced allergic airway inflammation by the induction of Th2 cytokines (35,36). However, nonallergic mechanisms by which IL19 and IL20 induce airway inflammation have been considered. Adenosine-induced IL19 production by primary bronchial epithelium cells enhanced monocyte TNFα production (37). In line with these literature data, we hypothesize that our findings underscore a central role of bronchial epithelial cells in the pathogenesis of recurrent wheeze after RSV LRTI.
In conclusion, genetic variation in adaptive immunity genes and particularly in IL10 family member genes IL19 and IL20 genes seems to be associated with recurrent wheeze after RSV LRTI, and perhaps infant wheeze in the general population, suggesting a role for IL19 and IL20 cytokines in airway disease. Investigations of how the IL19 and IL20 gene polymorphisms affect the function of the encoded protein or gene expression levels are needed to evaluate the pathophysiological mechanism underlying the protective effect of the TGG haplotype on recurrent wheeze after RSV LRTI.
Abbreviations
- FDR:
-
false discovery rate
- LD:
-
linkage disequilibrium
- LRTI:
-
lower respiratory tract infection
- RSV:
-
respiratory syncytial virus
- SNP:
-
single nucleotide polymorphism
References
Stein RT, Sherrill D, Morgan WJ, Holberg CJ, Halonen M, Taussig LM, Wright AL, Martinez FD 1999 Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. Lancet 354: 541–545
Bont L, van Aalderen WM, Versteegh J, Brus F, Draaisma JT, Pekelharing-Berghuis M, Van Diemen-Steenvoorde RA, Kimpen JL 2001 Airflow limitation during respiratory syncytial virus lower respiratory tract infection predicts recurrent wheezing. Pediatr Infect Dis J 20: 277–282
Ehlenfield DR, Cameron K, Welliver RC 2000 Eosinophilia at the time of respiratory syncytial virus bronchiolitis predicts childhood reactive airway disease. Pediatrics 105: 79–83
Bont L, Heijnen CJ, Kavelaars A, van Aalderen WM, Brus F, Draaisma JT, Geelen SM, Kimpen JL 2000 Monocyte IL-10 production during respiratory syncytial virus bronchiolitis is associated with recurrent wheezing in a one-year follow-up study. Am J Respir Crit Care Med 161: 1518–1523
Goetghebuer T, Isles K, Moore C, Thomson A, Kwiatkowski D, Hull J 2004 Genetic predisposition to wheeze following respiratory syncytial virus bronchiolitis. Clin Exp Allergy 34: 801–803
Ermers MJ, Hoebee B, Hodemaekers HM, Kimman TG, Kimpen JL, Bont L 2007 IL-13 genetic polymorphism identifies children with late wheezing after respiratory syncytial virus infection. J Allergy Clin Immunol 119: 1086–1091
Janssen R, Bont L, Siezen CL, Hodemaekers HM, Ermers MJ, Doornbos G, Van't Slot R, Wijmenga C, Houwelingen HC, Kimpen JL, Kimman TG, Hoebee B, Janssen R 2007 Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis 196: 826–834
Ermers MJ, Rovers MM, van Woensel JB, Kimpen JL, Bont LJ, RSV Corticosteroid Study Group 2009 The effect of high dose inhaled corticosteroids on wheeze in infants after respiratory syncytial virus infection: randomised double blind placebo controlled trial. BMJ 338: 897
Bont L, Steijn M, Van Aalderen WM, Brus F, Th Draaisma JM, Van Diemen-Steenvoorde RA, Pekelharing-Berghuis M, Kimpen JL 2004 Seasonality of long term wheezing following respiratory syncytial virus lower respiratory tract infection. Thorax 59: 512–516
Houben ML, Coenjaerts FE, Rossen JW, Belderbos ME, Hofland RW, Kimpen JL 2010 Disease severity and viral load are correlated in infants with primary respiratory syncytial virus infection in the community. J Med Virol 82: 1266–1271
Houben ML, Nikkels PG, van Bleek GM, Visser GH, Rovers MM, Kessel H, de Waal WJ, Schuijff L, Evers A, Kimpen JL, Bont L 2009 The association between intrauterine inflammation and spontaneous vaginal delivery at term: a cross-sectional study. PLoS ONE 4: e6572.
Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC 2004 A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20: 93–99
Dudbridge F 2008 Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered 66: 87–98
Benjamini Y, Hochberg Y 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300
Simões EA, Carbonell-Estrany X, Rieger CH, Mitchell I, Fredrick L, Groothuis JR, Palivizumab Long-Term Respiratory Outcomes Study Group 2010 The effect of respiratory syncytial virus on subsequent recurrent wheezing in atopic and nonatopic children. J Allergy Clin Immunol 126: 256–262
QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. Available at: http://hydra.usc.edu/gxe. Accessed March 30, 2011.
Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, Printz MC, Lee WM, Shult PA, Reisdorf E, Carlson-Dakes KT, Salazar LP, DaSilva DF, Tisler CJ, Gern JE, Lemanske RF Jr . 2008 Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med 178: 667–672
Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, Haugen H, Jelinek L, Kelly JD, Madden K, Maurer MF, Parrish-Novak J, Prunkard D, Sexson S, Sprecher C, Waggie K, West J, Whitmore TE, Yao L, Kuechle MK, Dale BA, Chandrasekher YA 2001 Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell 104: 9–19
Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, Kotenko SV 2000 Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes Immun 1: 442–450
Moore KW, de Waal MR, Coffman RL, O'Garra A 2001 Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19: 683–765
Bont L, Heijnen CJ, Kavelaars A, van Aalderen WM, Brus F, Draaisma JM, Pekelharing-Berghuis M, van Diemen-Steenvoorde RA, Kimpen JL 2001 Local interferon-gamma levels during respiratory syncytial virus lower respiratory tract infection are associated with disease severity. J Infect Dis 184: 355–358
Helminen M, Nuolivirta K, Virta M, Halkosalo A, Korppi M, Vesikari T, Hurme M 2008 IL-10 gene polymorphism at -1082 A/G is associated with severe rhinovirus bronchiolitis in infants. Pediatr Pulmonol 43: 391–395
Hoebee B, Bont L, Rietveld E, van Oosten M, Hodemaekers HM, Nagelkerke NJ, Neijens HJ, Kimpen JL, Kimman TG 2004 Influence of promoter variants of interleukin-10, interleukin-9, and tumor necrosis factor-alpha genes on respiratory syncytial virus bronchiolitis. J Infect Dis 189: 239–247
Wilson J, Rowlands K, Rockett K, Moore C, Lockhart E, Sharland M, Kwiatkowski D, Hull J 2005 Genetic variation at the IL10 gene locus is associated with severity of respiratory syncytial virus bronchiolitis. J Infect Dis 191: 1705–1709
Gentile DA, Doyle WJ, Zeevi A, Howe-Adams J, Kapadia S, Trecki J, Skoner DP 2003 Cytokine gene polymorphisms moderate illness severity in infants with respiratory syncytial virus infection. Hum Immunol 64: 338–344
Fickenscher H, Hor S, Kupers H, Knappe A, Wittmann S, Sticht H 2002 The interleukin-10 family of cytokines. Trends Immunol 23: 89–96
Dumoutier L, Leemans C, Lejeune D, Kotenko SV, Renauld JC 2001 Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J Immunol 167: 3545–3549
Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, Chandrasekher YA 2002 Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem 277: 47517–47523
Jordan WJ, Eskdale J, Boniotto M, Lennon GP, Peat J, Campbell JD, Gallagher G 2005 Human IL-19 regulates immunity through auto-induction of IL-19 and production of IL-10. Eur J Immunol 35: 1576–1582
Gallagher G, Eskdale J, Jordan W, Peat J, Campbell J, Boniotto M, Lennon GP, Dickensheets H, Donnelly RP 2004 Human interleukin-19 and its receptor: a potential role in the induction of Th2 responses. Int Immunopharmacol 4: 615–626
Oleksyk TK, Thio CL, Truelove AL, Goedert JJ, Donfield SM, Kirk GD, Thomas DL, O'Brien SJ, Smith MW 2005 Single nucleotide polymorphisms and haplotypes in the IL10 region associated with HCV clearance. Genes Immun 6: 347–357
Kõks S, Kingo K, Rätsep R, Karelson M, Silm H, Vasar E 2004 Combined haplotype analysis of the interleukin-19 and -20 genes: relationship to plaque-type psoriasis. Genes Immun 5: 662–667
Kingo K, Mossner R, Koks S, Ratsep R, Kruger U, Vasar E, Reich K, Silm H 2007 Association analysis of IL19, IL20 and IL24 genes in palmoplantar pustulosis. Br J Dermatol 156: 646–652
Fife MS, Gutierrez A, Ogilvie EM, Stock CJ, Samuel JM, Thomson W, Mack LF, Lewis CM, Woo P 2006 Novel IL10 gene family associations with systemic juvenile idiopathic arthritis. Arthritis Res Ther 8: R148.
Liao SC, Cheng YC, Wang YC, Wang CW, Yang SM, Yu CK, Shieh CC, Cheng KC, Lee MF, Chiang SR, Shieh JM, Chang MS 2004 IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol 173: 6712–6718
Huang F, Wachi S, Thai P, Loukoianov A, Tan KH, Forteza RM, Wu R 2008 Potentiation of IL-19 expression in airway epithelia by IL-17A and IL-4/IL-13: important implications in asthma. J Allergy Clin Immunol 121: 1415–1421
Zhong H, Wu Y, Belardinelli L, Zeng D 2006 A2B adenosine receptors induce IL-19 from bronchial epithelial cells, resulting in TNF-alpha increase. Am J Respir Cell Mol Biol 35: 587–592
Acknowledgements
We thank Jelle Goeman for providing his expertise on the global test for groups of genes and the RSV Corticosteroid Study Group for including patients.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by Dutch Asthma Foundation grant 3.2.03.22 and 3.2.07.001.
The authors report no conflicts of interest.
Rights and permissions
About this article
Cite this article
Ermers, M., Janssen, R., Onland-Moret, N. et al. IL10 Family Member Genes IL19 and IL20 Are Associated With Recurrent Wheeze After Respiratory Syncytial Virus Bronchiolitis. Pediatr Res 70, 518–523 (2011). https://doi.org/10.1203/PDR.0b013e31822f5863
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e31822f5863
This article is cited by
-
Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods
3 Biotech (2020)
-
The Burden and Long-term Respiratory Morbidity Associated with Respiratory Syncytial Virus Infection in Early Childhood
Infectious Diseases and Therapy (2017)
-
Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE)
BMC Pulmonary Medicine (2015)
-
Genetic polymorphisms and risk of recurrent wheezing in pediatric age
BMC Pulmonary Medicine (2014)
-
Leukemia inhibitory factor protects the lung during respiratory syncytial viral infection
BMC Immunology (2014)