Abstract
The pioneer microbiota of the neonate may affect future actions of the immune system. This study aimed to map the pioneer microbiota in healthy neonates vaginally born at term. A subgroup of neonates born large for GA (LGA) was compared with the neonates appropriate for GA (AGA). Fecal samples were collected, within 48 h after birth, from 79 neonates. Quantitative PCR was used for enumeration of Lactobacillus, a subgroup of Lactobacillus common in the vagina, Bifidobacterium, Enterococcus, Enterobacteriaceae, and the Bacteroides fragilis group. Cloning and sequencing were applied for subgroups of neonates born LGA or AGA. Lactobacillus was detected in all neonates, whereas other bacterial groups were detected only in 14 to 30% of the subjects. The prevalence of Gram-negative Proteobacteria was higher in neonates born LGA, whereas Gram-positive Firmicutes was more prevalent in neonates born AGA (p < 0.001). This study contributed to increased knowledge of the pioneer microbiota and indicates that neonates born LGA had significantly different microbiota compared with those born AGA. As the early microbiota can be important for maturation of the immune system, the outcome from this study may be relevant in the care of pregnant woman and newborns.
Similar content being viewed by others
Main
The fetal gastrointestinal tract is considered sterile, but at birth, the neonate is exposed to various bacteria from the birth canal via maternal vaginal and fecal microbiota, the hospital environment, and from handling by the parents and nursing staff (1). Various studies revealed immediate colonization of the newborn, the first days by only a few types of bacteria but a more complex ecosystem develops with time (2–4). The traditional view comprises initial colonization by facultative aerobes such as Enterobacteriaceae, Enterococcus, and Streptococcus (3,5) and later obligate anaerobes such as Bifidobacterium, Bacteroides, and Clostridium (5,6).
Microbes are thought to stimulate and tune the neonatal immune system and so the first microbiota can be of considerable importance to direct the neonatal immature immune system correctly (6–8). Lactobacillus is a genus of Gram-positive bacteria that dominates a healthy vagina (9). However, one third of women have bacterial vaginosis, which is characterized by reduced amount of lactobacilli, whereas other anaerobic bacteria, including Gram negatives, are more abundant (10,11). Gram-negative bacteria have lipopolysaccharides with strong proinflammatory capacity in the outer part of the cell wall, thereby contributing to a subclinical inflammatory tone connected to a range of metabolic disorders such as obesity and type 2 diabetes (12). Noteworthy, in neonates with a not yet fully mature immune system, lipopolysaccharide stimulation have different effects compared with adult individuals (13). Various reports show transfer of the maternal vaginal microbiota to the newborn (14,15), highlighting the importance of a healthy vaginal microbiota for desirable stimulation of the neonatal immune system.
Obese women and women with excessive weight gain during pregnancy more often give birth to neonates large for GA (LGA) (16). High birth weight is known to have impact on risk of metabolic disturbances, such as type 1 diabetes and increased BMI, later in life (17,18). Obesity is a risk factor for type 2 diabetes and has recently been linked to differences in intestinal microbiota (19,20), but how the neonatal microbiota affects the homeostasis later in life is unknown. The microbiota in neonates with normal birth weight has been studied [e.g. Palmer et al. (3)], but the intestinal ecosystem in neonates born LGA and the immediate and potentially lifelong influences needs further evaluation.
The present pilot study aimed, by direct gene identification and quantification, to map the first intestinal microbiota in healthy neonates vaginally born at term. A subgroup of neonates born LGA was compared with neonates born appropriate for GA (AGA). Fecal samples were collected within 48 h after birth and bacterial groups of interest were analyzed, as well as the dominating microbiota at different birth weights. To our knowledge, this is the largest study using molecular techniques on gut microbiota in the first days of life of neonates vaginally born at term.
METHODS
Subjects and sample collection.
Healthy singleton neonates vaginally born, without complications, at term were enrolled in this study, which was performed at an outpatient accommodation facility close to the labor ward at Skåne University Hospital, Malmö, Sweden, January through December 2008. Births with any complications, e.g. caesarean section, gestational diabetes, preterm delivery, or neonates born small for GA, remained in the maternity ward and were not recruited to this study. At the outpatient accommodation facility, parents and the newborn stay some days after delivery, and nursing staff give advice and recommendations about matters such as breastfeeding. In Sweden, all mothers are strictly advised to breastfeed if possible, and all participants presume to follow this guideline. Parents collected one fresh stool sample, the meconium, from the diaper within 48 h after birth, and samples were immediately frozen. The samples were regularly transported frozen to the laboratory and kept at −80°C until processing. The parents answered questionnaires about the pregnancy and delivery. Participation was voluntary, and parents gave written informed consent. The study was approved by the Regional Ethical Review Board in the south of Sweden.
DNA extraction.
Techniques based on amplification of the 16S rRNA gene require the bacteria to be lysed to facilitate DNA extraction. Consequently, DNA from stool content was isolated and purified by QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) in combination with glass bead beating to enhance disruption of bacterial cell walls, and BioRobot EZ1 (tissue kit and card; Qiagen) as described elsewhere (8). Negative controls for DNA extraction and downward analyses were run in parallel.
Quantitative PCR.
The amount of 16S rRNA genes of bacteria belonging to Lactobacillus, Bifidobacterium, Enterococcus, Enterobacteriaceae, and the Bacteroides fragilis group were estimated using separate qPCR assays. To elucidate whether vaginal lactobacilli, e.g. Lactobacillus crispatus and Lactobacillus gasseri were colonizing the neonates, a qPCR assay was designed with primers targeting the sequence of the 16S-23S rRNA intergenic spacer region [Lactobacillus group II; GII defined by Song et al. (21)]. For all assays, each reaction contained 10 μL 2× Rotor-Gene SYBR Green PCR Master Mix (Qiagen), 0.5 μM of each primer [Table 1, (21–27)], 2 μL of template DNA, and RNase-free water to the final volume of 20 μL. Samples, standards, and nontemplate controls were run in triplicate. The thermal cycling was performed in Rotor-Gene Q (Qiagen) with a program of 95°C for 5 min, followed by 40 cycles with denaturation at 95°C for 5 s, annealing, and elongation at 60°C for 10 to 30 s (Table 1). The fluorescent products were detected at the last step of each cycle. Melting curve analysis was made to ensure specific amplification. Absolute abundance of 16S rRNA genes was calculated based on standard curves using Rotor-Gene Q Series Software 1.7 (Qiagen), R2 >0.998. Detection limit was 104 genes/reaction for the B. fragilis group and Lactobacillus GII while all other assays detected 102 genes/reaction. As standard curves, cloned PCR products from Lactobacillus plantarum DSM9843, Lactobacillus iners CCUG28746T, Enterococcus faecalis CCUG19916T, Bifidobacterium infantis DSM15159, Escherichia coli CCUG29300T, and B. fragilis CCUG4856T were used. Ten-fold dilution series of the cloned PCR products were made in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) supplemented with 0.1 μg/μL herring sperm DNA (VWR International, West Chester, PA). Number of bacteria was expressed as numbers of 16S rRNA genes/g wet weight of feces.
Cloning and sequencing.
To investigate whether birth weight was related to the composition of the intestinal microbiota of newborns, the 16S rRNA genes from the feces of the five neonates with the highest birth weight, classified as LGA according to Marsál et al. (28), and five neonates with the lowest but still appropriate birth weight for GA, were cloned and sequenced. GA was 39 to 41½ wk. The 16S rRNA genes were amplified with the universal forward primer ENV1 (5′-AGA GTT TGA TII TGG CTC AG-3′) and the reverse primer ENV2 (5′-CGG ITA CCT TGT TAC GAC TT-3′), which anneal with 8–27 bp and 1511–1492 bp, respectively. PCR reaction mixture contained 0.2 μM of each primer, 0.2 mM of each deoxyribonucleotide triphosphate (Roche Diagnostics, Mannheim, Germany), 5 μL of 10 × PCR reaction buffer (100 mM Tris-HCl, 500 mM KCl, pH 8.3), 2 U/μL FastStart Taq polymerase (Roche Diagnostics), and 5 μL of template, in a final volume of 50 μL. Amplification was made for 30 cycles as described by Karlsson et al. (29), and PCR products were verified and purified as described elsewhere. Four PCR reactions from the same neonate were pooled and run on 1.5% agarose gel in Tris-acetate-EDTA (TAE) buffer. Bands were excised and DNA was purified with Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI). Purified PCR products were ligated into pGEM-T vector system and transformed into E. coli JM 109 according to the manufacturer's instructions (Promega). Colonies were blue/white screened on Luria-Bertani agar with Ampicillin (100 μg/mL; Sigma-Aldrich Co., St. Louis, MO), IPTG (0.5 mM, Promega), and X-Gal (80 μg/mL; Promega). Approximately 20 random white colonies were selected from each neonate and single-stranded sequenced by Eurofins MWG Operon (Ebersberg, Germany) using primer ENV1. Sequences were checked and edited in BioEdit Sequence Alignment Editor version 7.0.9.0. before submission to Ribosomal Database Project for phylogenetic affiliation to the closest database sequence with known identity.
Calculations.
Descriptive analyses were performed using SPSS Statistics 17.0.2 (SPSS, Inc., Chicago, IL). Data normally distributed were described with mean and SD whereas nonparametric data were described with median and 25th to 75th percentiles. For library comparison, the same number of sequences has to be used; therefore, 15 sequences from each neonate were randomly selected for overall clone library comparison with LIBSHUFF in mothur (30,31) and LibCompare with Naïve Bayesian rRNA Classifier, confidence threshold 80%, to reveal differences in microbial groups between neonates born LGA and AGA.
RESULTS
Cohort.
Seventy-nine neonates were enrolled in this study. All neonates were vaginally delivered, without complications, at gestational mean age of 40 wk and with a mean birth weight of 3682 g. Ten neonates were classified as LGA and the remaining 69 neonates were classified as AGA according to Marsál et al. (28). Between the two groups, there were no differences in GA or regarding antibiotic use during pregnancy. Characteristics of the cohort are summarized in Table 2.
Quantification of different bacterial groups.
Lactobacillus was detected with qPCR in all subjects (Fig. 1) with the load of 106 to 108 16S rRNA gene copies/g feces (Table 3). Bifidobacterium was found in 18% of the subjects, and Enterococcus was detectable in 27% but in relatively low numbers of the 16S rRNA gene. The Lactobacillus primers have previously been reported not to amplify enterococci (25), but we found the opposite (data not shown), so some detected lactobacilli could actually have been enterococci. However, the assay specific for Enterococcus only detected these in every third neonate, despite high assay sensitivity and specificity. The Gram-negative family Enterobacteriaceae was detected in 30% of the neonates, and the Gram-negative B. fragilis group in 14% (Fig. 1; Table 3). Bacteroides was found in strikingly high numbers per gram feces (Table 3). Eighteen percent of the neonates had detectable levels of Lactobacillus GII (Fig. 1; Table 3), which includes Lactobacillus species such as L. crispatus, L. gasseri, L. jensenii, and L. acidophilus (21). In this study, it was also observed that L. iners was amplified by the primers LU-1 and Lac-2. Notably, Lactobacillus GII was only detected in neonates born AGA. Bifidobacterium were more frequently detected in AGA, whereas 50% of neonates born LGA had Enterobacteriaceae and in fairly high numbers (Table 3).
Bacterial incidence in healthy neonates vaginally born at term. Incidence of different bacterial groups in the fecal microbiota of neonates in their first 48 h of life, presented as percentage of total number of neonates (n = 79). Primers used for the qPCR analysis are indicated in Table 1.
Comparison of the microbiota in neonates born LGA or AGA.
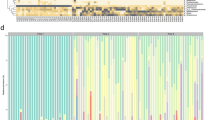
Cloning and sequencing were applied to further elucidate differences in neonates born LGA and AGA. Significant differences in the bacterial composition were found between neonates with high and appropriate birth weight (LIBSHUFF; p < 0.001). Significantly higher prevalence of sequences belonging to Proteobacteria was found in neonates born LGA (LibCompare; p < 0.001) and that was in accordance with the qPCR result (Table 3). Firmicutes were more prevalent in neonates born AGA (LibCompare; p < 0.001). No significant difference in Bacteroidetes was observed. Neonates were mainly dominated by a single genus, but the genus varied between individuals. The Gram-negative Escherichia was the most abundant genus in neonates born LGA, whereas Gram-positive genera such as Lactobacillus, Staphylococcus, and Clostridium were present among neonates born AGA. No difference in abundance of Enterococcus and Bacteroides was observed between the groups (Fig. 2). Sequences are deposited to GenBank, accession numbers HQ536093-HQ536167 for sequences from neonates born LGA and HQ536018-HQ536092 for neonates born AGA.
Microbial differences between neonates born LGA or AGA. Phylogenetic affiliation of sequences from neonates born LGA or AGA. Gender and birth weight (g) are indicated. Fifteen sequences from each neonate were used for clone library comparison. F, female; M, male. Escherichia  , Bacteroides
, Bacteroides  , Parabacteroides
, Parabacteroides  , Neisseria
, Neisseria  , Ralstonia
, Ralstonia  , Clostridium
, Clostridium  , Lactobacillus
, Lactobacillus  , Enterococcus
, Enterococcus  , Staphylococcus
, Staphylococcus  , Streptococcus
, Streptococcus  , Acidaminococcus
, Acidaminococcus  , Leuconostoc
, Leuconostoc  , and Bacillus
, and Bacillus  .
.
DISCUSSION
The pioneer microbiota of the entire cohort.
The neonatal gut microbiota may be important for later homeostasis (8). To our knowledge, this is the first report showing presence of lactobacilli in all neonates within the first 48 h of life. Previous studies report only sporadic lactobacilli presence at this early age (3,32). However, lactobacilli commonly dominate a healthy vagina (9,14) and have been found in breast milk (33,34). In this study, lactobacilli were detected in all neonates but Lactobacillus GII was only present in 20% of the AGA neonates and none of the LGA neonates had detectable levels. However, low sensitivity and target site discrepancy for the primer pairs may contribute to underestimation of Lactobacillus GII.
In this study, enterococci were detected in one third of the neonates. Enterococci are opportunistic pathogens causing, for example, nosocomial infections (35). Previous studies found these bacteria in two thirds of the subjects (36,37). However, those studies investigated the situation at the third and fourth day of life (36,37), which might have given the enterococci time to colonize and multiply to a higher degree. Enterococci can be contaminants from the environment at the outpatient accommodation facility and may not necessarily originate from the mother. When Enterococci were analyzed on the first day of life, similar colonization rates as the present study was obtained (34).
Diet influences the microbial ecosystem and breast milk is known to stimulate bifidobacteria and lactobacilli (34). Bifidobacterium is considered abundant in the neonatal microbiota (32), but in this cohort, only some of the neonates had detectable levels. However, previous studies have found Bifidobacterium in similar or even lower incidence at this early age (5,37). Detection discrepancy might be a consequence of different methods being used or differences in microbiota among mothers.
In one third of all neonates, Enterobacteriaceae was detected in the early microbiota. Adlerberth et al. (36) found E. coli in 40% of neonates at d 3, and 10% had other genera of Enterobacteriaceae. In a culture-based study from rural Guatemala, half of the neonates had E. coli during the first 24 h and all (n = 30) harbored this bacterium at their second day of life (5). Other investigations have found Enterobacteriaceae and E. coli in various amounts (2,3). Recently, E. coli in pregnant women was shown to positively correlate with neonate birth weight (38), which in turn, increases the risk of metabolic disturbances later in life (17,18). Moreover, offspring to rat dams supplemented with E. coli had higher Enterobacteriaceae load and increased gut permeability and systemic inflammation (39). The opposite was shown when rat dams were supplemented with the Gram-positive L. plantarum, suggesting altered physiology depending on the microbial transfer from mother to offspring (40). Interestingly, 50% of neonates born LGA were colonized by Enterobacteriaceae in substantial amounts. However, because of limited numbers of neonates born LGA, prevalence of the detected bacterial groups were too low to be statistically evaluated. The only exception would be Lactobacillus, but clearly there were no difference in prevalence and concentration of this bacterial group among neonates born LGA and AGA.
Only 14% of neonates in this cohort were colonized by strikingly high numbers of the Gram-negative obligate anaerobic B. fragilis group. In the literature, no clear evidence on B. fragilis colonization rate has been obtained at this early age.
The pioneer microbiota of subgroups of neonates born LGA or AGA.
Cloning and sequencing of the 16S rRNA genes were used to obtain a representation of the predominant microbiota. To the best of our knowledge, this is the first study among newborns with noncomplicated vaginal births to demonstrate that those born LGA have significantly different intestinal microbiota during their first 2 d of life compared with neonates born AGA. In accordance with detection of Enterobacteriaceae in neonates born LGA, these neonates harbored significantly more sequences corresponding to Gram-negative Proteobacteria, mainly E. coli, whereas neonates born AGA had significantly more sequences corresponding to Gram-positive Firmicutes and their microbiota seemed to be more diverse. Higher diversity has been reported in, for example, infants with atopic eczema compared with the ones without eczema (8) and diversity is further known to increase with age (4). Future studies should investigate how diversity differences early in life affect microbial ecosystem in adolescence and adulthood. Although cloning and sequencing were performed on only subgroups, the two methodological approaches used in this study resulted in similar results, namely that Proteobacteria was more prevalent in LGA. An explanation to the changed microbiota in LGA can be that these neonates may have obese mothers as maternal weight is known to affect infant microbiota (41).
Because the early microbiota can be important for maturation of the newborn immature immune system, the outcome from this study is of interest in the care of pregnant woman and newborns. Future studies should preferably address larger cohorts. They should elucidate the vaginal and the gastrointestinal microbiota of the mothers, the consequence of the pioneer microbiota on later homeostasis, and whether the microbial difference between neonates born LGA and AGA persist later in life.
Abbreviations
- AGA:
-
appropriate for GA
- LGA:
-
large for GA
References
Mackie RI, Sghir A, Gaskins HR 1999 Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69: 1035S–1045S
Songjinda P, Nakayama J, Kuroki Y, Tanaka S, Fukuda S, Kiyohara C, Yamamoto T, Izuchi K, Shirakawa T, Sonomoto K 2005 Molecular monitoring of the developmental bacterial community in the gastrointestinal tract of Japanese infants. Biosci Biotechnol Biochem 69: 638–641
Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO 2007 Development of the human infant intestinal microbiota. PLoS Biol 5: e177
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE 2011 Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108( suppl 1): 4578–4585
Mata LJ, Urrutia JJ, Lechtig A 1971 Infection and nutrition of children of a low socioeconomic rural community. Am J Clin Nutr 24: 249–259
Adlerberth I, Wold AE 2009 Establishment of the gut microbiota in Western infants. Acta Paediatr 98: 229–238
Levy O 2007 Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol 7: 379–390
Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Åberg N, Perkin MR, Tripodi S, Coates AR, Hesselmar B, Saalman R, Molin G, Ahrné S 2008 Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol 121: 129–134
Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G 2002 Vaginal Lactobacillus flora of healthy Swedish women. J Clin Microbiol 40: 2746–2749
Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH 2007 Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J Clin Microbiol 45: 1016–1018
Allsworth JE, Peipert JF 2007 Prevalence of bacterial vaginosis: 2001–2004 National Health and Nutrition Examination Survey data. Obstet Gynecol 109: 114–120
Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R 2007 Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56: 1761–1772
Nguyen M, Leuridan E, Zhang T, De Wit D, Willems F, Van Damme P, Goldman M, Goriely S 2010 Acquisition of adult-like TLR4 and TLR9 responses during the first year of life. PLoS ONE 5: e10407
Hegde S, Munshi AK 1998 Influence of the maternal vaginal microbiota on the oral microbiota of the newborn. J Clin Pediatr Dent 22: 317–321
Brook I, Barrett CT, Brinkman CR, Martin WJ, Finegold SM 1979 Aerobic and anaerobic bacterial flora of the maternal cervix and newborn gastric fluid and conjunctiva: a prospective study. Pediatrics 63: 451–455
Bodnar LM, Siega-Riz AM, Simhan HN, Himes KP, Abrams B 2010 Severe obesity, gestational weight gain, and adverse birth outcomes. Am J Clin Nutr 91: 1642–1648
Harder T, Roepke K, Diller N, Stechling Y, Dudenhausen JW, Plagemann A 2009 Birth weight, early weight gain, and subsequent risk of type 1 diabetes: systematic review and meta-analysis. Am J Epidemiol 169: 1428–1436
Gunnarsdottir I, Birgisdottir BE, Benediktsson R, Gudnason V, Thorsdottir I 2004 Association between size at birth, truncal fat and obesity in adult life and its contribution to blood pressure and coronary heart disease; study in a high birth weight population. Eur J Clin Nutr 58: 812–818
Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ 2008 Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 32: 1720–1724
Ley RE, Turnbaugh PJ, Klein S, Gordon JI 2006 Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023
Song Y, Kato N, Liu C, Matsumiya Y, Kato H, Watanabe K 2000 Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S–23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol Lett 187: 167–173
Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP 2001 Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 67: 2578–2585
Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM 2002 Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 68: 114–123
Malinen E, Rinttilä T, Kajander K, Matto J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A 2005 Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 100: 373–382
Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A 2004 Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 97: 1166–1177
Bartosch S, Fite A, Macfarlane GT, McMurdo ME 2004 Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol 70: 3575–3581
Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R 2004 Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol 70: 7220–7228
Marsál K, Persson PH, Larsen T, Lilja H, Selbing A, Sultan B 1996 Intrauterine growth curves based on ultrasonically estimated foetal weights. Acta Paediatr 85: 843–848
Karlsson C, Ahrné S, Molin G, Berggren A, Palmquist I, Fredrikson GN, Jeppsson B 2010 Probiotic therapy to men with incipient arteriosclerosis initiates increased bacterial diversity in colon: a randomized controlled trial. Atherosclerosis 208: 228–233
Singleton DR, Furlong MA, Rathbun SL, Whitman WB 2001 Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67: 4374–4376
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF 2009 Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541
Grönlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E 2000 Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0–6 months. Arch Dis Child Fetal Neonatal Ed 83: F186–F192
Martín R, Heilig GH, Zoetendal EG, Smidt H, Rodríguez JM 2007 Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol 103: 2638–2644
Solís G, de Los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M 2010 Establishment and development of lactic acid bacteria and Bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16: 307–310
Top J, Willems R, Bonten M 2008 Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol Med Microbiol 52: 297–308
Adlerberth I, Strachan DP, Matricardi PM, Ahrné S, Orfei L, Åberg N, Perkin MR, Tripodi S, Hesselmar B, Saalman R, Coates AR, Bonanno CL, Panetta V, Wold AE 2007 Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 120: 343–350
Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A 2008 Fecal microflora of Greek healthy neonates. Anaerobe 14: 94–101
Santacruz A, Collado MC, Garcia-Valdes L, Segura MT, Martin-Lagos JA, Anjos T, Marti-Romero M, Lopez RM, Florido J, Campoy C, Sanz Y 2010 Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 104: 83–92
Fåk F, Ahrné S, Molin G, Jeppsson B, Weström B 2008 Microbial manipulation of the rat dam changes bacterial colonization and alters properties of the gut in her offspring. Am J Physiol Gastrointest Liver Physiol 294: G148–G154
Fåk F, Ahrné S, Molin G, Jeppsson B, Weström B 2008 Maternal consumption of Lactobacillus plantarum 299v affects gastrointestinal growth and function in the suckling rat. Br J Nutr 100: 332–338
Collado MC, Isolauri E, Laitinen K, Salminen S 2010 Effect of mother's weight on infant's microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr 92: 1023–1030
Acknowledgements
We thank the staff at the outpatient accommodation facility (Egenvårdsavdelningen), Skåne University Hospital, and the parents and neonates for their contribution to this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the Swedish Research Council Formas, grant no. 222-2007-501, and The Crafoord Foundation.
Rights and permissions
About this article
Cite this article
Karlsson, C., Molin, G., Cilio, C. et al. The Pioneer Gut Microbiota in Human Neonates Vaginally Born at Term—A Pilot Study. Pediatr Res 70, 282–286 (2011). https://doi.org/10.1203/PDR.0b013e318225f765
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e318225f765
This article is cited by
-
Administration of probiotics to healthy volunteers: effects on reactivity of intestinal mucosa and systemic leukocytes
BMC Gastroenterology (2022)
-
Understanding the Connection Between the Gut–Brain Axis and Stress/Anxiety Disorders
Current Psychiatry Reports (2021)
-
Timing of complementary feeding is associated with gut microbiota diversity and composition and short chain fatty acid concentrations over the first year of life
BMC Microbiology (2020)
-
Enhanced nutrient supply and intestinal microbiota development in very low birth weight infants
Pediatric Research (2019)
-
Association between birth route and late-onset sepsis in very preterm neonates
Journal of Perinatology (2016)