Abstract
Ureaplasma spp. is detected in the urogenital tract, including the vagina, cervix, chorioamnion, and placenta. Their colonization is associated with histologic chorioamnionitis (CAM), often observed in placentas from preterm delivery. We isolated Ureaplasma spp. from 63 preterm placentas among 151 specimens, which were delivered at <32 wk of gestation. Of the 63 placentas, 52 (83%) revealed CAM in cultures positive for Ureaplasma spp., however, CAM was observed only in 30% (26/88) of cultures negative for Ureaplasma spp. (p < 0.01). Colonization by Ureaplasma spp. was an independent risk factor for CAM (OR, 11.27; 95% CI, 5.09–24.98). Characteristic neutrophil infiltration was observed in the amnion and subchorion (bistratified pattern) in cultures positive for Ureaplasma spp. FISH analysis of CAM placenta with male infant pregnancy indicated that bistratified infiltrated neutrophils showed the XX karyotype and umbilical vein infiltrated neutrophils showed XY karyotype. The distribution of sulfoglycolipid, the receptor of Ureaplasma spp., was mainly detected in the amnion. Ureaplasmal urease D protein and ureB gene were both detected in the amnion, indicating direct colonization by Ureaplasma spp.
Similar content being viewed by others
Main
Ureaplasma spp. is the smallest self-replicating organism, both in genome size and in cellular dimensions. It lacks cell walls and exists in association with eukaryotic cells, mainly colonizing mucosal surfaces of the respiratory and urogenital tracts (1). Ureaplasma spp. is a common inhabitant of the lower genital tract and isolated from 40 to 80% women of child-bearing age (2). However, once Ureaplasma spp. spreads from the lower genital tract into the body, this microorganism exerts widespread pathogenic effects, such as chorioamnionitis (CAM), urinary tract infections, preterm labor, and spontaneous abortion. On the other hand, Ureaplasma spp. infection is also reported as a risk factor for lethal pneumonia, chronic lung disease, and meningitis of fetuses and neonates (3).
CAM is a placental finding associated with premature rupture of membranes (PROM) and preterm birth, which are the most important causes of perinatal morbidity and mortality (4,5). Previous studies showed that CAM was positively related to the isolation of Ureaplasma spp (6,7). Although many researchers reported the detection of Ureaplasma spp. from specimens of vagina, cervix, chorioamnion, and placenta using culture or PCR methods (8–12), the precise pathologic findings of CAM with Ureaplasma spp. remain unclear.
A variety of infectious microorganisms use specific host cell surface molecules as receptors. Such receptors provide a mechanism for intimate interaction with the host cell membrane and in some cases may facilitate the subsequent entry of the organism into the cell (13). Ureaplasma spp. and Mycoplasma hominis were shown to specifically recognize host cell surface glycolipids (sulfogalactoglycerolipid and the sphingolipid counterpart, sulfogalactosyl ceramide), which have been implicated in sperm-egg interactions (14). This glycolipid receptor binding may relate to the reproductive pathogenesis of these organisms. Furthermore, there are no previous reports about the specific receptor of Ureaplasma spp. and its distribution in the placenta.
This study was conducted to confirm the prevalence of placental Ureaplasma spp. in preterm delivery, and whether there is an association between Ureaplasma spp. and CAM. Moreover, we identified placental features that might be characteristic of ureaplasmal infection.
METHODS
Subjects and placental examination.
All clinical specimens were obtained after informed consent approved by the Ethics Committee of Osaka Medical Center and Research Institute for Maternal and Child Health. In this study from January to December 2007, pathologic examinations of 151 placentas, including 67 cesarean deliveries, delivered at <32 wk of gestation were performed. As a control, 41 term placentas [mean gestational age, 39.6 wk (SD 1.1); mean birth weight, 3009 g (SD 425)], including 11 cesarean deliveries (27%), four PROM (10%), and two intrauterine growth retardation (5%), were also examined. The pathologists who examined the placentas were blinded to the results of Ureaplasma spp. culture. Placentas were examined according to the method of Fox and Sebire (15). The histologic criterion used for CAM was the presence of accumulated leukocytes extending through the fetal membranes using Blanc's classification (16). Umbilical vasculitis was defined as migration of fetal inflammatory cells into or through the media of the umbilical arteries or veins. The criterion used for subacute necrotizing funisitis was a typical deposition of calcification around the vessels in the umbilical cord.
Culture for the detection of Ureaplasma spp.
A microbiologist who was blinded to all clinical details of the patients prepared cultures for the detection of Ureaplasma spp.; species identification and subtyping of Ureaplasma spp. were also developed. The placental swabs were collected from the fetal side of the placentas. These were suspended in UMCHs medium: Mycoplasma broth base (Becton, Dickinson and Co., Baltimore, MD) 1.47% (wt/vol), yeast extract (Becton, Dickinson and Co.) 2.5% (wt/vol), horse serum (Biowhittaker, Walkersville, MD) 20% (vol/vol), supplement VX (Becton, Dickinson and Co.) 1.0% (vol/vol), urea 0.04% (wt/vol), phenol red 0.001% (wt/vol), l-cysteine hydrochloride 0.01% (wt/vol), and penicillin G 1000 U/mL. After incubation at 35°C for up to 72 h, the color of the medium changed from yellow to red due to hydrolysis of urea, and these color changes were regarded as indicating positivity for Ureaplasma spp. We identified Ureaplasma spp. by colony formation and subsequent PCR-based assays using modified Kong's method (17).
Immunohistochemistry.
Paraffin-embedded sections were deparaffinized and rehydrated. The sections were incubated with rabbit anti-human myeloperoxidase polyclonal antibody (1:300; Dako, Tokyo, Japan), mouse anti-human cluster of differentiation (CD) 45 MAb (1:1; Thermo/Shandon Immunon, Pittsburgh, PA), mouse anti-human CD68 MAb (1:50; Dako), chicken anti-Ureaplasma urealyticum UreD polyclonal antibody (1:100; Abcam, Cambridge, MA), and mouse anti-sulfoglycolipid MAb (1:5) (18). Immunoreactivity was detected using the Envision Kit (Dako) with horseradish peroxidase-conjugated anti-chicken IgG antibody (1:200; Bethyl Laboratories, Montgomery, TX). Sections were counterstained with Mayer's hematoxylin (Muto Pure Chemicals, Tokyo, Japan).
FISH analysis.
X/Y FISH was performed using the CEP X/Y DNA Probe Kit (Vysis, Downers Grove, IL) according to the manufacturer's instructions. Included in this kit are probes for Xp11.1-q11.1 of chromosome X (labeled with Spectrum Red) and for Yq12 of chromosome Y (labeled with Spectrum Green). Fluorescence microscope (Olympus IX71 microscope; Olympus, Tokyo, Japan) integrated with a digitized CCD camera (Nikon Digital Sight DS-5Mc, Nikon Instech, Kanagawa, Japan) and imaging software (Nikon ACT-2U Nikon) were used to investigate and analyze the FISH results with DAPI II counterstain.
PCR for the detection of Ureaplasma urease structural gene.
DNA was extracted from paraffin-embedded placental sections using DNA Isolator PS-Rapid Reagent (Wako Pure Chemical Industries, Osaka, Japan) according to the manufacturer's instructions. The DNA solution was used for subsequent PCRs. The PCR primers used were as follows: Glut1, sense (5′-TGCAAGGGGAAAGGAAAAGG-3′) and antisense (5′-GAAGAGAAACTCTGCCCTGC-3′) of the human facilitative glucose transporter genes; and ureB, sense (5′-CCAGGTAA ATTAGTACCAGG-3′) and antisense (5′-CCTGATGGAATATCGAAACG-3′) of the Ureaplasma urease structural genes. Twenty-five micro liters of GoTaq Green Master Mix (Promega, Madison, WI), 1 μL sample, and 14 μL water were added to each reaction. A thermal cycler was used to process samples through 35 cycles at 95°C for 30 s, 64°C for 30 s, and 68°C for 30 s for Glut1 and through 35 cycles at 95°C for 30 s, 45°C for 30 s, and 68°C for 30 s for ureB.
Statistical analysis.
All statistical analyses were performed using SPSS 11.0 for Windows (SPSS, Chicago, IL). The χ2 test or Fisher's exact test was used to compare the incidence of each placental feature between cases culture positive and negative for Ureaplasma spp. Logistic regression models were used to examine the association between culture positivity for Ureaplasma spp. and the severity of CAM. The Mann-Whitney U test or unpaired t test was used to compare continuous variables. A difference was considered significant when the p value was <0.05. Logistic regression analysis, adjusting for the potential confounding clinical factors associated with CAM, were conducted to evaluate the independent association of Ureaplasma spp. colonization with CAM. The strength of association in these models is reported as the adjusted OR with the 95% CI.
RESULTS
Incidence of Ureaplasma spp. colonization in preterm placentas.
Among 151 placentas delivered at <32 wk of gestation, 42% (63/151) were culture positive for Ureaplasma spp. Figure 1A shows the incidence of Ureaplasma spp. colonization in preterm placentas according to gestational age. Forty-nine percent (38/77) of placentas delivered during the second trimester (at 13–26 wk) and 34% (25/74) delivered during the early third trimester (at 27–31 wk) were colonized with Ureaplasma spp. The incidence of Ureaplasma spp. colonization during the second trimester was higher than that during the early third trimester, but the difference was not statistically significant (49% versus 34%; p = 0.05). The incidence of Ureaplasma spp. colonization in term placentas was 24% (10/41); this was significantly lower than that in preterm placentas (24% versus 42%; p < 0.05). On the contrary, 33% (22/67) of preterm cesarean delivery placentas (at 21–31 wk) were culture positive for Ureaplasma spp. (Fig. 1B).
Isolation of Ureaplasma spp. from preterm placenta. Forty-two percent (63/151) of preterm placentas (<32 wk) were culture positive for Ureaplasma spp. (A), and 33% (22/67) in preterm cesarean delivery, (B) black bars, culture positive for Ureaplasma spp.; white bars, culture negative for Ureaplasma spp.
Maternal characteristics associated with Ureaplasma spp. colonization.
Maternal characteristics of the Ureaplasma-positive and -negative groups are shown in Table 1. The incidence of PROM in the Ureaplasma-positive group was significantly higher than that in the negative group (44% versus 16%; p < 0.01). The incidences of pre-eclampsia and cesarean section in the Ureaplasma-positive group were lower than those in the negative group (5% versus 17%, p < 0.05; 35% versus 51%, p < 0.05, respectively). There were no significant differences between these two groups in other maternal characteristics. Other microorganisms cultured from placentas are listed in Table 2. Among 151 placentas, only 13 (9%) were culture positive for other microorganisms, the most frequent of which were group B Streptococcus (GBS) and Escherichia coli.
Pathologic features of placentas in Ureaplasma-positive group.
Placental findings in the Ureaplasma-positive and -negative groups are presented in Table 3. Placentas of the Ureaplasma-positive group showed a significantly higher frequency of CAM compared with the negative group (83% versus 30%; p < 0.01). CAM with umbilical cord inflammation was also more frequent in the Ureaplasma-positive group than in the negative group (46% versus 7%; p < 0.01). No differences were found in the percentages of any other findings between these two groups.
Association of Ureaplasma spp. colonization with CAM.
Table 4 displays ORs for the strength of association between histologic CAM and risk factors for the development of CAM. Culture positivity for Ureaplasma spp. and PROM were significantly associated with CAM (OR, 11.27; 95% CI, 5.09–24.98; and OR, 7.67; 95% CI, 3.13–18.84, respectively). The associations between culture positivity for Ureaplasma spp. and CAM and between PROM and CAM remained significant after adjustment for confounding factors in logistic regression analyses (OR, 8.60; 95% CI, 3.72–19.89; and OR, 5.36; 95% CI, 1.98–14.52, respectively).
Characteristics of CAM placentas colonized with Ureaplasma spp.
To explore the characteristics of preterm CAM placentas colonized with Ureaplasma spp., we compared 52 CAM placentas in the Ureaplasma-positive group with 26 CAM placentas in the negative group (Table 5). Logistic regression models using Blanc I CAM as the reference showed that culture positivity for Ureaplasma spp. was associated with severe CAM (Blanc II and III).
Moreover, we found a characteristic pathologic finding named the “amniotic bistratified pattern,” in which a stratified leukocyte infiltration in the amnion and subchorion was accompanied by necrosis of the amniotic epithelium (Fig. 2A), in contrast to GBS positive (control) placenta (Fig. 2B). This pattern was significantly more frequent in the Ureaplasma-positive group than in the negative group (35% versus 8%; p < 0.05). It counted 51% (18/35) of Blanc III CAM in the Ureaplasma-positive group. In all 20 placentas that showed the specific neutrophils infiltration of amniotic bistratified pattern, 13 placentas were colonized with Ureaplasma spp. solely, five placentas with both Ureaplasma spp. and other microorganisms (Escherichia coli, Pseudomonas fluorescens, Enterobacter cloacae, Candida glabrata, Streptococcus mitis, or Clostridium spp.), and two placentas without any microorganisms (Table 2). None of the placentas, which were negative for Ureaplasma spp. and positive for other microorganisms showed this amniotic bistratified placental pattern.
Amniotic bistratified neutrophil infiltration in placenta with CAM. A, C–F, Amniotic bistratified pattern in Blanc III CAM placenta colonized with Ureaplasma spp. B, Blanc III CAM placenta colonized with GBS. A–C, Mayer's Hematoxylin and eosin (H&E) stain. D, Myeloperoxidase. E, CD45. F, CD68. Arrows indicate infiltrated leukocytes.
Specific placental features of intrauterine Ureaplasma infection.
To determine the characteristics of the bistratified infiltrated cells in the amnion and subchorionic space, we analyzed Ureaplasma-positive Blanc III CAM placentas and GBS-positive (control) placentas by immunohistochemical staining. Most of the bistratified infiltrated cells in Ureaplasma-positive placentas were myeloperoxidase-positive neutrophils, indicating acute inflammation, whereas immunohistochemical staining of CD45 and CD68 were negative (Fig. 2C–F). Control CAM placentas with GBS infection also showed acute inflammation (data not shown).
Next, we analyzed the origin of the migrated neutrophils showing bistratified patterning around the Ureaplasma-positive placental membrane (Fig. 3A). Unexpectedly, infiltrated inflammatory cells in the amnion and the subchorionic space showed only two red signals (XX genotype), indicating maternally derived cells (Fig. 3B and C). On the other hand, trophoblasts in the villi and inflammatory cells migrating through the wall of the umbilical vessels (Fig. 3E and F) showed one red and one green signals (fetal XY genotype), as expected (Fig. 3D and G).
X(red)/Y(green) FISH analysis. (A) H&E stain around the membrane. X/Y FISH of (B) amnion, (C) subchorionic space, and (D) villi. (E) H&E stain of umbilical cord. (F) Higher magnification of umbilical vein and infiltrated cells. (G) X/Y FISH of infiltrated cells. One green and one red signal indicate XY karyotype (male, fetus-derived cells) and two red signals indicate XX karyotype (female, maternal-derived cells). DAPI II was used as a counterstain.
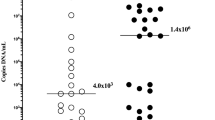
To explain these maternal immunologic responses, we further analyzed the localization of the microorganism's DNA in the Ureaplasma-positive placenta. PCR amplification of the Ureaplasma-specific ureB gene was detected only in purified genomic DNA from the chorioamnion (Fig. 4A, B-1) and not detected in villi (Fig. 4A, B-2) or decidua (Fig. 4A, B-3) in the paraffin-embedded sections that showed bistratified infiltration of neutrophils (Fig. 4B, upper column). On the contrary, PCR amplification of human Glut1 gene was detected in genomic DNA obtained from chorioamnion, villi, and decidua (Fig. 4B, lower column). By immunohistochemical staining with anti-Ureaplasma UreD antibody, which was a polyclonal antibody against the Ureaplasma urease, positive signals were detected in the amnion where maternal neutrophils had infiltrated (Fig. 4C and D). Immunohistochemical staining of sulfoglycolipid, the receptor of Ureaplasma spp., showed that it was mainly distributed in the amnion in normal term placenta (Fig. 4E).
Distribution of Ureaplasma spp. and its receptor sulfoglycolipid in placenta. A, H&E stain. B, PCR for the detection of Ureaplasma DNA. (Upper column) The Ureaplasma ureB gene is detected in genomic DNA from chorioamnion (lane 1) but not in villi and decidua (lanes 2 and 3, respectively). PC, positive control (full Ureaplasma spp. genome). NC, negative control (distilled water). (Lower column) The human Glut1 gene is detected in genomic DNA from chorioamnion, villi, and decidua (lanes 1, 2 and 3, respectively). PC, positive control (full human genome). NC, negative control (distilled water). C, D, Ureaplasma UreD is detected in the amnion where maternal neutrophils had infiltrated. E, Sulfoglycolipid is mainly distributed in the amnion in normal term placenta.
DISCUSSION
Infectious and inflammatory processes in the uterus during pregnancy remain a major cause of preterm delivery and subsequent complications in newborn infants. Pathologic CAM is frequently associated with preterm delivery. The most common microbes isolated from the amniotic cavity of women with preterm labor are Ureaplasma spp. and Mycoplasma hominis (19). Studies based on the isolation of Ureaplasma spp. from the placenta uniformly showed a significant association with CAM (9,10). The stimulatory effect of Ureaplasma spp. on cytokine release, such as tumor necrosis factor-α, IL-8, and IL-6, has been confirmed in vitro (20–22). Multiple-banded antigen and other lipoproteins from Ureaplasma spp. were found to activate nuclear factor kappaB through Toll-like receptor (TLR) 1, TLR2, and TLR6 and induce tumor necrosis factor-α in mouse peritoneal macrophages (23), indicating that not only viable Ureaplasma spp. but its lipoproteins cause an excessive immune response in utero. Umbilical vasculitis and chorionic plate inflammation might be caused by these antigenic components from local infection. To confirm the association of Ureaplasma spp. with CAM, we conducted Ureaplasma spp. cultivation as a prospective cohort study of 151 preterm placentas. Furthermore, immunohistochemical, PCR, and FISH analyses were performed in the infected placentas to confirm the microbial localizations.
We examined the incidence of Ureaplasma spp. colonization in the placenta by using the culture method. Studies involving various clinical specimens have shown PCR to be more sensitive than conventional culture methods (24–26). However, given the high sensitivity of PCR, a false positive result for a particular organism is more likely to occur than when culture methods are used, principally because of intersample contamination. Furthermore, PCR detects nonviable organisms as well as viable organisms, and does not enable further analysis using isolated microorganisms. On the other hand, culture for detection of Ureaplasma spp. is expensive and requires specialized media and expertise that are not widely available. We therefore established a culture method for the detection of Ureaplasma spp. by modification of the method of Shepard and Lunceford (27) Shepard and Combs (27,28), which uses materials that are all commercially available and easily prepared just before use.
The relation between infection and preterm delivery is not consistent throughout gestation. Infection is rare in late preterm delivery but is present in most cases in which birth occurs at <30 wk (29,30). Previous evidence suggests that intrauterine infection may occur early in pregnancy. For example, Ureaplasma spp. has been detected in some samples of amniotic fluid obtained in routine chromosomal analysis at 15–18 wk of gestation. In most of these women, delivery was around 24 wk (31–33). Meanwhile, Perni et al. (34) reported that in 179 asymptomatic women who received amniocentesis at midtrimester, all women with preterm PROM (5/5) tested positive for either Ureaplasma spp. or Mycoplasma hominis as opposed to none of the women (0/5) with spontaneous preterm birth. In this study, we showed that the incidence of Ureaplasma spp. colonization in preterm placentas was statistically higher than that in term placentas (p < 0.05) and that the incidence during the second trimester was higher than that during early in the third trimester (p = 0.05). We suggest that Ureaplasma infection-related CAM might be the leading cause of preterm delivery during the second trimester. One possible explanation for these gestational age-related changes in the cause of preterm delivery is that the intrauterine immune system during the second trimester might be susceptible to weakly pathogenic microorganisms such as Ureaplasma spp., and the hormonal immune response or cytokine production necessary to initiate labor is easily activated. In our study, the culture positive for other microorganism (Table 2) was lower (13/151; 8.6%) than those in previous reports (35,36). This discrepancy might be caused by 1) the treatment for inflammation and/or infection by bacteriocidal agents, which reduced the colonization in placentas, and 2) the samples, which we used was the placental surface swab but not by the homogenized tissues.
Although the association between Ureaplasma spp. colonization and CAM has already been identified, details of the placental features remain unclear. We therefore examined in detail all placentas delivered at <32 wk of gestation and investigated the presence of features specific to Ureaplasma spp. colonization. In the CAM placentas, polymorphonuclear leukocytes first accumulate in the intervillous space immediately below the chorionic plate, which forms the roof of this space. The inflammatory cells in the roof of the intervillous space later extend upward into the chorionic plate and reach the amnion. At this stage, the inflammatory cellular response is purely maternal in origin, the leukocytes being derived from maternal blood in the intervillous space (15). According to this migration process, we can simply imagine that this accumulation is composed of one layer of inflammatory cells extending toward the amniotic cavity. However, we found a characteristic pathologic feature named the amniotic bistratified pattern in which leukocytes did not infiltrate in one layer but in two layers. This pathologic pattern was significantly more frequent in placentas colonized with Ureaplasma spp. than that in others. Therefore, this amniotic bistratified pattern might be a specific placental feature of intrauterine Ureaplasma infection.
To explain how this infiltration pattern develops, we first hypothesized that inflammatory cells in the subchorionic space are derived from maternal sources, but those in the amnion are derived from fetal sources. However, we showed that neutrophils in both the subchorionic space and amnion were maternal cells. On the contrary, infiltrated neutrophils in the umbilical vein were of fetal origin (Fig. 3D and G). Earlier findings might be due to the differences in production and response between maternal and fetal neutrophils. Immature newborn infants frequently become neutropenic (37) in their response to bacterial sepsis. However, adults develop a sustained neutrophil leukocytosis by releasing preformed neutrophils from the marrow storage pool into circulation and increasing proliferation by recruiting a great number of committed granulocyte progenitors into the cell cycle. Furthermore, the most consistently observed functional abnormality of neonatal neutrophils is reduced chemotaxis. In most assays, neutrophils from newborn infants migrate at about half the speed of adult cells (38). Neonatal neutrophils display less interaction with endothelial monolayers in conditions of flow than adult cells. Rolling adhesion is diminished, and fewer cells attach to activated endothelium and migrate to the subendothelial tissue (39). These reports suggest that maternal neutrophils react predominantly at the fetomaternal interface; on the contrary, fetal neutrophils possess the immunologic reaction at least in the fetal side.
Next, we hypothesized that the development of this pattern might be involved in the distribution of genomic DNA and protein from Ureaplasma spp. as well as its receptor sulfoglycolipid in the placenta. There are no previous reports of their distribution, but we demonstrated that the urease structural gene and protein of Ureaplasma spp. were distributed in the amnion where maternal neutrophils accumulated and that their receptor sulfoglycolipid was also present in the amnion. Therefore, we suggest that this characteristic pattern of maternal neutrophil infiltration in the amnion might be associated with the distribution of Ureaplasma spp. and its receptor in the placenta.
In conclusion, our study indicates the following: 1) ∼40–50% of preterm placentas delivered at <32 wk of gestation are culture positive for Ureaplasma spp.; 2) placentas colonized with Ureaplasma spp. show CAM significantly more frequently than others; 3) positivity of cultures for Ureaplasma spp. is an independent risk factor for CAM; 4) positivity of cultures for Ureaplasma spp. is also associated with the severity of CAM; and 5) the amniotic bistratified pattern in CAM might be a specific placental feature for intrauterine Ureaplasma infection.
Abbreviations
- CAM:
-
chorioamnionitis
- GBS:
-
group B Streptococcus
- PROM:
-
premature rupture of membranes
- TLR:
-
toll-like receptor
References
Waites KB, Schelonka RL, Xiao L, Grigsby PL, Novy MJ 2009 Congenital and opportunistic infections: Ureaplasma species and Mycoplasma hominis. Semin Fetal Neonatal Med 14: 190–199
Taylor-Robinson D, McCormack WM 1980 The genital mycoplasmas. N Engl J Med 302: 1003–1010
Cassell GH, Waites KB, Watson HL, Crouse DT, Harasawa R 1993 Ureaplasma urealyticun intrauterine infection: role in prematurity and disease in newborns. Clin Microbiol Rev 6: 69–87
Goldenberg RL 2002 The management of premature labor. Obstet Gynecol 100: 1020–1037
Wood NS, Marlow N, Costeloe K, Gibson AT, Wilkinson AR 2000 Neurologic and developmental disability after extremely preterm birth. EPICure Study Group. N Engl J Med 343: 378–384
Goldenberg RL, Rouse DJ 1998 Prevention of preterm birth. N Engl J Med 339: 313–320
Shurin PA, Alpert S, Bernard Rosner BA, Driscoll SG, Lee YH 1975 Chorioamnionitis and colonization of the newborn infant with genital mycoplasmas. N Engl J Med 293: 5–8
Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, Eschenbach DA 1988 A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N Engl J Med 319: 972–978
Kundsin RB, Driscoll SG, Monson RR, Yeh C, Biano SA, Cochran WD 1984 Association of Ureaplasma urealyticum in the placenta with perinatal morbidity and mortality. N Engl J Med 310: 941–945
Embree JE, Krause VW, Embil JA, MacDonald S 1980 Placental infection with Mycoplasma hominis and Ureaplasma urealyticum: clinical correlation. Obstet Gynecol 56: 475–481
Honma Y, Yada Y, Takahashi N, Momoi MY, Nakamura Y 2007 Certain type of chronic lung disease of newborns is associated with Ureaplasma urealyticum infection in utero. Pediatr Int 49: 479–484
Naessens A, Foulon W, Cammu H, Goossens A, Lauwers S 1987 Epidemiology and pathogenesis of Ureaplasma urealyticum in spontaneous abortion and early preterm labor. Acta Obstet Gynecol Scand 66: 513–516
Revesz T, Greaves M 1975 Ligand-induced redistribution of lymphocyte membrane ganglioside GM1. Nature 257: 103–106
Lingwood CA, Quinn PA, Wilansky S, Nutikka A, Ruhnke HL, Miller RB 1990 Common sulfoglycolipid receptor for mycoplasmas involved in animal and human infertility. Biol Reprod 43: 694–697
Fox H, Sebire NJ 2007 Pathology of the Placenta. Saunders Elsevier, Philadelphia
Blanc WA 1981 Pathology of the placenta, membranes, and umbilical cord in bacterial, fungal and viral infections in man. Monogr Pathol 67–132
Kong F, Zhu X, Wang W, Zhou X, Gordon S, Gilbert GL 1999 Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1. J Clin Microbiol 37: 538–543
Cheng X, Zhang Y, Kotani N, Watanabe T, Lee S, Wang X, Kawashima I, Tai T, Taniguchi N, Honke K 2005 Production of a recombinant single-chain variable-fragment (scFv) antibody against sulfoglycolipid. J Biochem 137: 415–421
Romero R, Mazor M 1988 Infection and preterm labor. Clin Obstet Gynecol 31: 553–584
Manimtim WM, Hasday JD, Hester L, Fairchild KD, Lovchik JC, Viscardi RM 2001 Ureaplasma urealyticum modulates endotoxin-induced cytokine release by human monocytes derived from preterm and term newborns and adults. Infect Immun 69: 3906–3915
Li YH, Brauner A, Jonsson B, van der Ploeg I, Soder O, Holst M, Jensen JS, Lagercrantz H, Tullus K 2000 Ureaplasma urealyticum-induced production of proinflammatory cytokines by macrophages. Pediatr Res 48: 114–119
Viscardi RM, Hasday JD 2009 Role of Ureaplasma species in neonatal chronic lung disease: epidemiologic and experimental evidence. Pediatr Res 65: 84R–90R
Shimizu T, Kida Y, Kuwano K 2008 Ureaplasma parvum lipoproteins, including MB antigen, activate NF-kappaB through TLR1, TLR2 and TLR6. Microbiology 154: 1318–1325
Teng K, Li M, Yu W, Li H, Shen D, Liu D 1994 Comparison of PCR with culture for detection of Ureaplasma urealyticum in clinical samples from patients with urogenital infections. J Clin Microbiol 32: 2232–2234
Abele-Horn M, Wolff C, Dressel P, Zimmermann A, Vahlensieck W, Pfaff F, Ruckdeschel G 1996 Polymerase chain reaction versus culture for detection of Ureaplasma urealyticum and Mycoplasma hominis in the urogenital tract of adults and the respiratory tract of newborns. Eur J Clin Microbiol Infect Dis 15: 595–598
Cunliffe NA, Fergusson S, Davidson F, Lyon A, Ross PW 1996 Comparison of culture with the polymerase chain reaction for detection of Ureaplasma urealyticum in endotracheal aspirates of preterm infants. J Med Microbiol 45: 27–30
Shepard MC, Lunceford CD 1978 Serological typing of Ureaplasma urealyticum isolates from urethritis patients by an agar growth inhibition method. J Clin Microbiol 8: 566–574
Shepard MC, Combs RS 1979 Enhancement of Ureaplasma urealyticum growth on a differential agar medium (A7B) by a polyamine, putrescine. J Clin Microbiol 10: 931–933
Watts DH, Krohn MA, Hillier SL, Eschenbach DA 1992 The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet Gynecol 79: 351–357
Chellam VG, Rushton DI 1985 Chorioamnionitis and funiculitis in the placentas of 200 births weighing less than 2.5 kg. Br J Obstet Gynaecol 92: 808–814
Gray DJ, Robinson HB, Malone J, Thomson RB Jr 1992 Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat Diagn 12: 111–117
Horowitz S, Mazor M, Romero R, Horowitz J, Glezerman M 1995 Infection of the amniotic cavity with Ureaplasma urealyticum in the midtrimester of pregnancy. J Reprod Med 40: 375–379
Nguyen DP, Gerber S, Hohlfeld P, Sandrine G, Witkin SS 2004 Mycoplasma hominis in mid-trimester amniotic fluid: relation to pregnancy outcome. J Perinat Med 32: 323–326
Perni SC, Vardhana S, Korneeva I, Tuttle SL, Paraskevas LR, Chasen ST, Kalish RB, Witkin SS 2004 Mycoplasma hominis and Ureaplasma urealyticum in midtrimester amniotic fluid: association with amniotic fluid cytokine levels and pregnancy outcome. Am J Obstet Gynecol 191: 1382–1386
Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, Extremely Low Gestational Age Newborns (ELGAN) Study Investigators 2008 Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am J Obstet Gynecol 198: 110.e1–110.e7
Svensson L, Ingemarsson I, Mårdh PA 1986 Chorioamnionitis and the isolation of microorganisms from the placenta. Obstet Gynecol 67: 403–409
Christensen RD 1989 Neutrophil kinetics in the fetus and neonate. Am J Pediatr Hematol Oncol 11: 215–223
Wolach B, Sonnenschein D, Gavrieli R, Chomsky O, Pomeranz A, Yuli I 1998 Neonatal neutrophil inflammatory responses: parallel studies of light scattering, cell polarization, chemotaxis, superoxide release, and bactericidal activity. Am J Hematol 58: 8–15
Anderson DC, Abbassi O, Kishimoto TK, Koenig JM, Mclntire LV, Smith CW 1991 Diminished lectin-, epidermal growth factor-, compliment binding domain-cell adhesion molecule-1 on neonatal neutrophils underlies their impaired CD18-independent adhesion to endothelial cells in vitro. J Immunol 146: 3372–3379
Acknowledgements
We thank Drs. Koichi Honke, Akihiro Morita, Naohiro Yonemoto, and Atsushi Tabata for their helpful advices, and Futoshi Fujiwara, Keiko Matsuoka, Yuko Kuwae, and Akiko Koide for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by grants-in-aid from the Ministry of Health, Labor and Welfare, Japan; Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; Research on Child Health and Development, Japan; and The Foundation for Mother and Child Well-being, Osaka, Japan.
Rights and permissions
About this article
Cite this article
Namba, F., Hasegawa, T., Nakayama, M. et al. Placental Features of Chorioamnionitis Colonized With Ureaplasma Species in Preterm Delivery. Pediatr Res 67, 166–172 (2010). https://doi.org/10.1203/PDR.0b013e3181c6e58e
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e3181c6e58e
This article is cited by
-
Ureaplasma parvum infection induces inflammatory changes in vaginal epithelial cells independent of sialidase
Molecular Biology Reports (2023)
-
Vaginal Ureaplasma species increase chorioamnionitis in very preterm infants with preterm premature rupture of the membranes at < 28 weeks of gestation
European Journal of Clinical Microbiology & Infectious Diseases (2018)
-
Human thioredoxin-1 attenuates the rate of lipopolysaccharide-induced preterm delivery in mice in association with its anti-inflammatory effect
Pediatric Research (2016)