Abstract
Rhinovirus (RV), a member of the Picornaviridae family, accounts for many virus-induced asthma exacerbations. RV induces airway cell chemokine expression both in vivo and in vitro. Because of the known interactions of proteases with cellular functions, we hypothesized that RV 3C protease is sufficient for cytokine up-regulation. A cDNA encoding RV16 3C protease was constructed by PCR amplification and transfected into 16HBE14o− human bronchial epithelial cells. 3C protease induced expression of both IL-8 and GM-CSF, as well as transcription from both the IL-8 and GM-CSF promoters. 3C expression also induced activator protein 1 and NF-κB transcriptional activation. Finally, mutation of IL-8 promoter AP-1 and NF-κB promoter sequences significantly reduced 3C-induced responses. Together, these data suggest expression of RV16 3C protease is sufficient to induce chemokine expression in human bronchial epithelial cells, and does so in an AP-1- and NF-κB–dependent manner.
Similar content being viewed by others
Main
RVs are the most common upper respiratory pathogens, inducing the majority of common colds worldwide. Viral infections trigger asthma in 80 to 85% of asthma exacerbations in children and 44% in adults (1, 2) RV, a member of the Picornaviridae family of small, positive-stranded RNA viruses, accounts for much of virus-induced asthma exacerbations (1, 2). Cytokines are elaborated in vivo in the nasal mucosa, and most likely also from bronchial epithelium, after RV infection. Neutrophils, eosinophils, and lymphocytes increase in the nasal secretions of RV-infected patients compared with uninfected controls (3, 4).
The cytokines IL-8 and GM-CSF have been implicated in the pathogenesis of asthma. IL-8 is a potent activator and chemoattractant of neutrophils (5) and eosinophils (6), and GM-CSF promotes the differentiation and survival of recruited eosinophils (7). Increased levels of IL-8, GM-CSF (8), and ICAM-1 (9), the receptor of 90% of RV serotypes, as well as IL-1 and IL-6, have been found in the nasal secretions of individuals infected with RVs (10–12). In bronchial mucosal biopsies obtained after experimental infection with RV, Bardin and colleagues (13) noted a significant increase in submucosal lymphocytes and epithelial eosinophils, which, in asthmatics, persisted into convalescence. Although increases in cytokine secretion in the nasal mucosa does not prove increased expression in the epithelium in the lower airways, taken together the above data strongly suggest that cytokine elaboration is increased in the RV-infected lower airway epithelium.
RV infection also induces cytokine production and adhesion molecule expression in cell culture. Divergent findings regarding cytokine and adhesion molecule stimulation have arisen from studies using different RV serotypes and different cell types. RV-induced IL-6 and IL-8 production has been noted in MRC-5 fibroblasts and A549, BEAS-2B, and primary bronchial epithelial cells (8, 12, 14, 15). GM-CSF expression is up-regulated in response to RV infection in BEAS-2B bronchial epithelial cells (8). VCAM-1 expression is increased by RV infection in 16HBE14o− cells (9). IL-6, IL-8, and ICAM-1 elaboration in response to RV infection occurs at the level of transcription (9, 12, 14).
The picornavirus genome encodes a 5′ and 3′ noncoding region and a single polyprotein that is secondarily cleaved into mature proteins and 5′ and 3′ noncoding regions. The polyprotein is divided into three regions; P1, P2, and P3. P1 is cleaved into four mature capsid proteins. P2 and P3 are cleaved into seven mature nonstructural proteins; of these, 2A and 3C are proteases (16). 2A plays a prominent role in shut-off of host protein synthesis after viral infection (17), and 3C performs most of the cleavages of the viral polyprotein. Because of the known interactions of the proteases with cellular functions, we hypothesized that expression of the 3C protease is sufficient for cytokine up-regulation in airway epithelial cells. We found that RV16 3C protease was sufficient to increase IL-8 and GM-CSF expression in 16HBE14o− human bronchial epithelial cells. As far as we are aware, this is the first published report demonstrating that an individual RV protein up-regulates cytokine responses. We also demonstrate that 3C-induced IL-8 transcription was dependent on AP-1 and NF-κB promoter sequences.
METHODS
Cell culture.
A derivative of 16HBE14o− human bronchial epithelial cells, provided by S. White (University of Chicago, Chicago, IL, U.S.A.), was studied. Cell lines were originally established from bronchial epithelial tissue by transfection with pSVori−, which contains the origin-defective SV40 genome (18). Unlike the parental line, these cells do not grow in distinct clusters and demonstrate improved transfection efficiency. Cultures show specific immunostaining with pan-cytokeratin c11 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), bind galactose- or galactosamine-specific lectins particular to basal epithelial cells (19), and express β1-, α2-, α3-, and α6-integrin subunits on their cell surface (20). Cells were grown on coated plates (fibronectin, 10 μg/mL; collagen, 30 μg/mL; BSA, 100 μg/mL). 16HBE14o− and HeLa cells were grown in Dulbecco's modified eagle medium with 10% fetal bovine serum, glutamine, nonessential amino acids, amphotericin B, and gentamicin as described previously (21). Use of a human bronchial cell line was approved by the University of Chicago Institutional Review Board.
Plasmids.
pCMV3C was constructed by PCR amplification of the 3C gene in the pRV1611.1 infectious clone kindly provided by W.M. Lee (University of Wisconsin, Madison, WI, U.S.A.). Primers for PCR amplification encoded the complementary sequence to that at the boundaries of 3C determined by Lee et al.(22). The PCR product was ligated with the eukaryotic TOPO TA cloning vector (encoding a CMV promoter upstream of the PCR insert) according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, U.S.A.). Reporter constructs encoding a 206-bp fragment of the human IL-8 promoter (−162 to +44) and mutations of the IL-8 promoter NF-κB and AP-1 sites (ΔNF-κB −162/+44 hIL-8/Luc and ΔAP-1 −162/+44 hIL-8/Luc) upstream of the luciferase gene were kindly provided by A.R. Brasier (University of Texas Medical Branch, Galveston, U.S.A.) (23, 24). The GM-CSF reporter construct (pGMluc37) was kindly provided by P.N. Cockerill (Institute for Medical and Veterinary Science, Adelaide, Australia), and encoded a 655-bp fragment of the human GM-CSF promoter (−627 to +28) upstream of the luciferase gene (25). pCMVβ-gal encodes the CMV promoter upstream of the β-galactosidase gene (Promega Corporation, Madison, WI, U.S.A.). NF-κB-TATA/Luc and AP-1-TATA/Luc reporter plasmids were purchased from Stratagene (La Jolla, CA, U.S.A.).
3C mRNA detection.
16HBE14o− cells were transfected with pCMV3C or empty vector in a six-well plate. Twenty-four hours after transfection, the cells were washed twice with PBS and lysed directly in the plate by adding 1 mL of TriZol Reagent (Invitrogen). Total RNA was isolated according to the manufacturer's instructions. RNA was treated with RNase-free DNase I at 37°C for 30 min to degrade any trace amounts of genomic or plasmid DNA, followed by a 5-min incubation at 75°C and phenol-chloroform extraction. Reverse transcription was performed for 60 min at 42°C using a 3C- or human glyceraldehyde-3-phosphate dehydrogenase (GADPH)–specific reverse primer. PCR amplification of cDNA was performed in a total volume of 25 μL with the primers 3C forward (caggatatcgccaccatgGGTCCAGAAGAAGAATT) and 3C reverse (caggcggccgcctaggcgtagtcgggcacgtcgtaggggtaTTGTTGTTCAGTGAAG). As an internal control, human GAPDH was amplified from the same RNA samples as those from which the 3C gene was amplified. The PCR conditions were 40 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min. A 610-bp product was visualized by ethidium bromide staining after electrophoresis on a 2% agarose gel.
IL-8 and GM-CSF protein quantification.
Immunoreactive IL-8 and GM-CSF in the supernatants of cultured 16HBE14o− cells were quantified by dual-antibody ELISA. Antibodies were obtained from Endogen (Cambridge, MA, U.S.A.).
Reporter assays.
16HBE14o− cells were cotransfected with either pCMV3C or empty vector and either −162hIL-8/Luc, pGMluc37, NF-κB-TATA/Luc, AP-1-TATA/Luc, ΔNF-κB −162/+44 hIL-8/Luc or ΔAP-1 −162/+44 hIL-8/Luc. Lipofectamine (Life Technologies, Inc., Rockville, MD, U.S.A.) was used as previously described (26). 16HBE14o− cells were seeded into six-well plates, and 4 μL of lipofectamine was added to each well. Selected cultures were treated with TNF-α (2–10 ng/mL for 16 h).
Statistical analysis.
The data were expressed as mean ± SEM. For reporter assays, changes in promoter activity were calculated as arbitrary light units/β-galactosidase calorimetric units per hour. Group mean comparisons between empty vector and RV16 3C were performed with an unpaired t test.
RESULTS
Expression of 3C protein increases production of IL-8 and GM-CSF proteins.
RV proteins may directly interact with cellular factors to stimulate cytokine production. To test this hypothesis, 16HBE14o− cells were transfected with a cDNA clone (pCMV3C) encoding a CMV promoter upstream of the gene encoding one RV16 protein, the 3C protease. 3C mRNA was detected by reverse transcriptase PCR in pCMV3C-transfected 16HBE14o− cell lysate 24 h after transfection, indicating that 3C mRNA was expressed (Fig. 1).
Detection of 3C mRNA by reverse transcription (RT)-PCR of 16HBE14o− cell lysate harvested 24 h after transfection with pCMV3C. PCR product of pCMV3C (lanes 1, 2, and 3) and empty vector negative control (lanes 4, 5, and 6) transfected cells are visualized on an ethidium bromide–stained 2% agarose gel. As an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also amplified and compared. To remove contaminating DNA, and its possible amplification from RNA preparation, total RNA was divided into three groups: first, DNaseI-treated and -reverse transcribed (lanes 1 and 4); second, DNaseI-treated but not reverse transcribed (lanes 2 and 5); third, without any treatment (lanes 3 and 6).
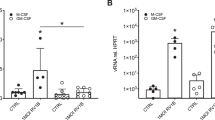
After transfection, supernatant was removed for measurement of IL-8 and GM-CSF protein by ELISA. Despite a transfection efficiency of 10 to 20%, IL-8 protein abundance increased 2- to 3-fold after transfection of pCMV3C, relative to cells transfected with empty vector (Fig. 2A). In addition, there was a 2.5-fold increase in GM-CSF protein in the supernatant of pCMV3C-transfected cultures compared with the empty vector control (Fig. 2B).
RV16 3C increases transcription from the IL-8 and GM-CSF promoters.
RV-induced up-regulation of IL-6, IL-8, and ICAM-1 occurs at the level of transcription in A549 or BEAS-2B cells (9, 12, 14). To determine whether RV 3C-induced IL-8 and GM-CSF response in 16HBE14o− cells is transcriptionally regulated, 16HBE14o− cells were transfected with IL-8 (−162hIL-8/Luc) or GM-CSF (pGMluc37) promoter constructs expressing the luciferase reporter gene. Cells were cotransfected with pCMV3C or empty vector and pCMVβ-gal. After 48 h, cells were harvested and luciferase and β-galactosidase activities were measured, and promoter activity normalized for transfection efficiency. 3C expression significantly increased IL-8 promoter activity relative to empty vector control (p < 0.05;Fig. 3A). The GM-CSF promoter activity increased nearly 3-fold over the empty vector negative control and approached the increase seen after TNF-α treatment (p < 0.05;Fig. 3B).
Effects of RV16 3C protein expression on transcription from the IL-8 and GM-CSF promoters. 16HBE14o− cells were transfected with pCMV3C, pCMVβ-gal and either −162hIL-8/Luc (A) or pGMluc37 (B) 48 h before cell harvest. For comparison, selected cultures were treated with TNF-α (10 ng/mL). Promoter activity represents luciferase activity measured in light units normalized for transfection efficiency with β-galactosidase activity measured in OD (n = 4–8, *p < 0.05 relative to empty vector).
RV16 3C protease up-regulation of IL-8 promoter activity is AP-1– and NF-κB–dependent.
We have determined that IL-8 expression in 16HBE14o− cells is dependent on the transcriptional activity of both AP-1 and NF-κB promoter sequences (27). To determine whether RV16 3C induced AP-1 and NF-κB transactivation, 16HBE14o− cells were transfected with reporter constructs encoding the sequences of either the NF-κB or AP-1 binding site upstream of a TATA box and the luciferase gene. Expression of 3C significantly increased AP-1 transcriptional activity relative to cells transfected with empty vector, to a level similar to TNF-α (Fig. 4A). Transfection with pCMV3C also significantly increased NF-κB transactivation, although the level was substantially less than that elicited by TNF-α treatment (Figs. 4B).
To determine the requirements of the IL-8 promoter NF-κB and AP-1 binding sites, cells were cotransfected with RV16 3C protease or empty vector and IL-8 reporter plasmids with mutated NF-κB or AP-1 sites (ΔNF-κB −162/+44 hIL-8/Luc or ΔAP-1 −162/+44 hIL-8/Luc). Mutation of the NF-κB site reduced the basal level of promoter activity, as well as 3C and TNF-α responsiveness (Fig. 5). Mutation of the AP-1 site attenuated the basal level of activity and 3C-induced transcription from the IL-8 promoter. Mutation of the AP-1 site also attenuated the absolute level of TNF-α–induced responses, but the fold increase relative to the basal level was unchanged, consistent with the idea that the AP-1 site serves as a basal level enhancer in the context of TNF-α stimulation. Taken together, these data suggest that RV16 3C protease up-regulation of IL-8 promoter activity is AP-1– and NF-κB–dependent.
DISCUSSION
RV stimulates IL-8 and GM-CSF production in A549, MRC-5, and BEAS-2B cells (8, 14, 15, 28). Little is known about the events initiating RV-induced cytokine expression in airway epithelium. The IL-8 response to RV serotype 16 infection in BEAS-2B human respiratory epithelial cells is inhibited by antibodies to ICAM-1, implying that integrin-mediated signal transduction pathways play a role in RV-induced responses (29). On the other hand, in primary bronchial epithelial cells, anti–ICAM-1 attenuates RANTES but not IL-8 secretion (30). After ICAM-1 binding, RV14 requires an active endocytic pathway for successful infection of HeLa cervical adenocarcinoma cells (31), consistent with the notion that endocytosis-dependent signaling pathways may also be required for RV16-induced epithelial cell responses. Finally, it is conceivable that specific RV components, either the RNA itself or encoded viral proteins, are necessary for epithelial cell cytokine and ICAM-1 expression. Because of the known interactions of the proteases with cellular functions, we hypothesized that expression of the 3C protease is sufficient for cytokine up-regulation in airway epithelial cells. We therefore tested the sufficiency of RV16 3C protease for human bronchial epithelial cell chemokine production. We demonstrate that the RV16 3C protease is sufficient for IL-8 and GM-CSF protein expression. This increase in chemokine expression occurs at the level of transcription, because the IL-8 and GM-CSF promoters were stimulated in response to the 3C protein. The RV16 3C protease increased AP-1 and NF-κB transactivation, and the 3C-induced transcription from the IL-8 promoter is dependent on both AP-1 and NF-κB promoter sequences.
These data extend the work of Zalman and colleagues (32), who examined the requirement of 3C protease for airway epithelial cell responses. In BEAS-2B cells, treatment with AG7088, an inhibitor of 3C protease, inhibited the production of IL-6 and IL-8. Together with our data, this study indicates that 3C is both required and sufficient for IL-8 expression. We are aware of one other instance in which an individual viral protein has been shown to enhance transcription of IL-8. The adenoviral protein E1A protein promotes cell entry into the S phase of the cell cycle, facilitating viral replication. Gilmour et al.(33) found that stable transfection of A549 cells with E1A up-regulated IL-8 protein elaboration induced by treatment with lipopolysaccharide or environment particulate matter <10 μM. As in the current study, the sufficiency of the viral protein in question was tested by transfecting respiratory epithelial cells with a plasmid expressing one viral protein (in this case E1A). This being the case, one cannot be certain in either instance that overexpression of viral protein did not play a role in the observed results.
Although expression of RV16 3C protease was sufficient for 16HBE14o− bronchial epithelial cell IL-8 expression, it is conceivable that the up-regulation of IL-8 by the virus itself is the outcome of multiple influences of individual RV proteins or RNA acting in an additive or synergistic manner. For example, poliovirus 3A has been shown to down-regulate IL-8 elaboration in human MG63 fibrosarcoma cells (34). Down-regulation of the RV-induced cytokine response from one of the other RV16 proteins could serve to attenuate the impact of 3C. As noted above, effects of virus attachment (29) and entry (31) could also play a role.
There are conflicting reports regarding the NF-κB dependence of RV-induced cytokine and adhesion molecule up-regulation. Zhu et al.(12, 14) reported that RV14 stimulation of IL-6 and IL-8 in MRC-5 and A549 cells is NF-κB dependent. Mutation of the NF-κB sites in the IL-6 and IL-8 promoters decreased both basal and RV14-induced IL-6 and IL-8 responses. Papi and Johnston (9) showed in A549 cells that RV16-induced ICAM-1 promoter activity is dependent on binding of NF-κB proteins to the −187 to −178 NF-κB binding site on the ICAM-1 promoter in A549 cells. Papi and Johnston (35) also showed in A549 and 16HBE14o− cells that RV16-induced VCAM-1 promoter activity is dependent on protein binding to NF-κB binding sites in the promoter. However, Kim et al.(29) found that RV induction of IL-6 and IL-8 in BEAS-2B cells is NF-κB independent. In the latter report, sulfasalazine and calpain inhibitor 1, chemical inhibitors of NF-κB activation, attenuated NF-κB binding but not RV16-induced IL-6 or IL-8 mRNA expression in BEAS-2B cells. These authors also found that RV16-induced GM-CSF production was blocked by sulfasalazine, suggesting that in this system, GM-CSF expression, but not that of IL-6 or IL-8, was NF-κB dependent. Relatively little information is available regarding the role of AP-1 promoter sequences in RV-mediated responses. Papi and Johnston (35) showed that deletion of a proximal AP-1 site had no effect on RV16-induced VCAM-1 promoter activity. In the current study, we found that RV16 3C induced both AP-1 and NF-κB transactivation. Further, mutation of the IL-8 promoter AP-1 and NF-κB sites reduced 3C responsiveness, suggesting that RV16 3C protease-induced transcription from the IL-8 promoter is AP-1 and NF-κB dependent. Differences between our results and the aforementioned earlier studies may be caused by differences in serotype or viral strain. Also, the signaling pathways to chemokine in 16HBE14o− cells expressing RV16 3C may be different from those used after infection by whole RV.
We did not determine the mechanism by which RV16 3C protease regulates AP-1 and NF-κB transcriptional activity. It has been shown in HeLa cells that RV16 3C enters the nucleus and cleaves Oct-1 and transcription factor II D transcription factors (34), consistent with the notion that 3C may enhance transcription by directly interacting with components of the basal transcription complex as well as upstream transcription factors. Hepatitis B pX, which has been demonstrated to transactivate viral and cellular genes under the control of AP-1 and NF-κB and interact with components of the basal transcription complex, also induces NF-κB activation independently of (IKB kinase) (36). On the other hand, 3C protease may also induce chemokine expression by initiating upstream signaling pathways.
CONCLUSIONS
In summary, we have shown that expression of RV16 3C protease in 16HBE14o− human bronchial epithelial cells induces expression of IL-8 and GM-CSF via AP-1– and NF-κB–dependent pathways. Investigation of potential pathways to RV-induced cytokine up-regulation including the 3C-initiated pathway will enhance the understanding of RV-induced inflammatory responses.
Abbreviations
- AP:
-
activator protein
- E1A:
-
early region 1A
- GM-CSF:
-
granulocyte-macrophage colony-stimulating factor
- ICAM-1:
-
intercellular adhesion molecule-1
- NF:
-
nuclear factor
- RV:
-
rhinovirus
- TNF-α:
-
tumor necrosis factor-α
- VCAM:
-
vascular cell adhesion molecule
References
Nicholson KG, Kent J, Ireland DC 1993 Respiratory viruses and exacerbations of asthma in adults. BMJ 307: 982–986
Johnston SL, Pattemore G, Sanderson S, Smith F, Lampe L, Josephs P, Symington S, O'Toole SH, Myint DA, Tyrrell AJ, Holgate ST 1995 Community study of role of virus infections in exacerbations of asthma in 9–11 year old children. BMJ 310: 1225–1228
Heymann PW, Rakes AD, Ingram GE, Hoover GE, Platts-Mills TAE 1995 Assessment of eosinophils, viruses, and IgE antibody in wheezing infants and children. Int Arch Allergy Immunol 107: 380–382
Levandowski RA, Weaver CW, Jackson GG 1988 Nasal-secretion leukocyte populations determined by flow cytometry during acute rhinovirus infection. J Med Virol 25: 423–432
Mills P, Davies R, Devalia J 1999 Airway epithelial cells, cytokines, and pollutants. Am J Respir Crit Care Med 160( suppl): S38–S43
Sehmi R, Cromwell O, Wardlaw A, Moqbel R, Kay A 1993 Interleukin-8 is a chemo-attractant for eosinophils purified from subjects with a blood eosinophilia but not from normal healthy subjects. Clin Exp Allergy 23: 1027–1036
Borish L, Rosenwasser L 1996 Update on cytokines. J Allergy Clin Immunol 97: 719–734
Subauste MC, Jacoby DB, Richards SM, Proud D 1995 Infection of a human respiratory epithelial cell line with rhinovirus: induction of cytokine release and modulation of susceptibility to infection by cytokine exposure. J Clin Invest 96: 549–557
Papi A, Johnston SL 1999 Rhinovirus infection induces expression of its own receptor intercellular adhesion molecule 1 (ICAM-1) via increased NF-κB-mediated transcription. J Biol Chem 274: 9707–9720
Grunberg K, Timmers MC, Smits HH, de Klerk EPA, Dick EC, Spaan WJM, Hiemstra PS, Sterk PJ 1997 Effect of experimental rhinovirus 16 colds on airway hyperresponsiveness to histamine and interleukin-8 in nasal lavage in asthmatic subjects in vivo. Clin Exp Allergy 27: 36–45
Proud D, Gwaltney JJ, Hendley J, Dinarello C, Gillis S, Schleimer R 1994 Increased levels of interleukin-1 are detected in nasal secretions of volunteers during experimental rhinovirus colds. J Infect Dis 169: 1007–1013
Zhu Z, Tang W, Ray A Y W, Einarsson O, Landry ML, Gwaltney J, Elias JA 1996 Rhinovirus stimulation of interleukin-6 in vivo and in vitro: evidence for nuclear factor kappa B-dependent transcriptional activation. J Clin Invest 97: 421–430
Bardin PG, Fraenkel DJ, Sandeson G, Lampe F, Holgate ST 1995 Lower airways inflammatory response during rhinovirus colds. Int Arch Allergy Immunol 107: 127–129
Zhu Z, Tang W, Wu Y, Elias JA 1997 Rhinovirus stimulation of interleukin-8 in vivo and in vitro: role of NF-κB. Am J Physiol 273: L814–L824
Kaul P, Biagioli MC, Singh I, Turner RB 2000 Rhinovirus-induced oxidative stress and interleukin-8 elaboration involves p47-phox but is independent of attachment to intercellular adhesion molecule-1 and viral replication. J Infect Dis 181: 1885–1890
Alvey J, Wyckoff E, Yu S, Lloyd R, Ehrenfeld E 1991 cis- and trans- cleavage activities of poliovirus 2A protease expressed in Escherichia coli. J Virol 65: 6077–6083
Barco A, Feduchi E, Carrasco L 2000 A stable HeLa cell line that inducibly expresses poliovirus 2A(pro): effects on cellular and viral gene expression. J Virol 74: 2383–2392
Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC 1994 CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol 10: 38–47
Dorscheid DR, Conforti AE, Hamann KJ, Rabe KF, White SR 1999 Characterization of cell surface lectin-binding patterns of human airway epithelium. Histochem J 31: 145–151
White SR, Dorscheid DR, Rabe KF, Wojcik KR, Hamann KJ 1999 Role of very late adhesion integrins in mediating repair of human airway epithelial cell monolayers after mechanical injury. Am J Respir Cell Mol Biol 20: 787–796
Funkhouser A, Purcell R, D'Hondt E, Emerson S 1994 Attenuated hepatitis A virus: genetic determinants of adaptation to growth in MRC-5 cells. J Virol 68: 148–157
Lee W-M, Wang W, Rueckert R 1994 Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9: 177–181
Garofalo R, Sabry M, Jamaluddin M, Yu RK, Casola A, Ogra PL, Brasier AR 1996 Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J Virol 70: 8773–8781
Brasier AR, Jamaluddin M, Casola A, Duan W, Shen Q, Garofalo RP 1998 A promoter recruitment mechanism for tumor necrosis factor-alpha-induced interleukin-8 transcription in type II pulmonary epithelial cells: dependence on nuclear abundance of Rel A, NF-kappaB1, and c-Rel transcription factors. J Biol Chem 273: 3551–3556
Cockerill G, Bert A, Ryan G, Gamble J, Vadas M, Cockerill P 1995 Regulation of granulocyte-macrophage colony-stimulating factor and E-selection expression in endothelial cells by cyclosporin A and the T-cell transcription factor NFAT. Blood 86: 2689–2698
Page K, Li J, Hodge JA, Liu PT, Vanden Hoek TL, Becker LB, Pestell RG, Rosner MR, Hershenson MB 1999 Characterization of a Rac1 signaling pathway to cyclin D1 expression in airway smooth muscle cells. J Biol Chem 274: 22065–22071
Li J, Kartha S, Tan A, Iasvovskaia S, Bhat RK, Manaligod JM, Page K, Brasier AR, Hershenson MB 2002 Regulation of human airway epithelial cell interleukin-8 expression by mitogen-activated protein kinases. Am J Physiol Lung Cell Mol Physiol 283: L690–L699
Sanders SP, Siekierski ES, Porter JD, Richards SM, Proud D 1998 Nitric oxide inhibits rhinovirus-induced cytokine production and viral replication in a human respiratory epithelial cell line. J Virol 72: 934–942
Kim J, Sanders S, Siekierski E, Casolaro V, Proud D 2000 Role of NF-kappa B in cytokine production induced from human airway epithelial cells by rhinovirus infection. J Immunol 165: 3384–3392
Schroth MK, Grimm E, Frindt P, Galagan DM, Konno S-I, Love R, Gern JE 1999 Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol 20: 1220–1228
DeTulleo L, Kirchhausen T 1998 The clathrin endocytic pathway in viral infection. EMBO J 17: 4585–4593
Zalman LS, Brothers MA, Dragovich PS, Zhou R, Prins TJ, Worland ST, Patick AK 2000 Inhibition of human rhinovirus-induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother 44: 1236–1241
Gilmour PS, Rahman I, Hayashi S, Hogg JC, Donaldson K, MacNee W 2001 Adenoviral E1A primes alveolar epithelial cells to PM10-induced transcription of interleukin-8. Am J Physiol Lung Cell Mol Physiol 281: L598–L606
Dodd DA, Giddings TH, Kirkegaard K 2001 Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J Virol 75: 8158–8165
Papi A, Johnston SL 1999 Respiratory epithelial cell expression of vascular cell adhesion molecule-1 and its up-regulation by rhinovirus infection via NF-κB and GATA transcription factors. J Biol Chem 274: 30041–30051
Purcell NH, Yu C, He D, Xiang J, Paran N, DiDonato JA, Yamaoka S, Shaul Y, Lin A 2001 Activation of NF-κB by hepatitis B virus X protein through an IκB kinase-independent mechanism. Am J Physiol Gastrointest Liver Physiol 280: G669–G677
Acknowledgements
The authors thank W.M. Lee for his gift of the pRV1611.1 infectious clone, S. White for providing 16HBE14o− cells, and A.R. Brasier and P.N. Cockerill for providing reporter plasmids.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by National Institutes of Health Grant HL56399.A.W.F. and J.A.K. contributed equally to the work.
Rights and permissions
About this article
Cite this article
Funkhouser, A., Kang, JA., Tan, A. et al. Rhinovirus 16 3C Protease Induces Interleukin-8 and Granulocyte-Macrophage Colony-Stimulating Factor Expression in Human Bronchial Epithelial Cells. Pediatr Res 55, 13–18 (2004). https://doi.org/10.1203/01.PDR.0000099801.06360.AB
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/01.PDR.0000099801.06360.AB